Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells
- PMID: 34624216
- PMCID: PMC8555837
- DOI: 10.1016/j.molcel.2021.09.013
Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells
Abstract
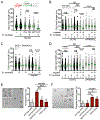
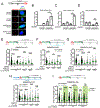
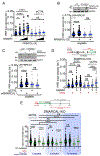
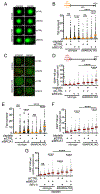
PRIMPOL repriming allows DNA replication to skip DNA lesions, leading to ssDNA gaps. These gaps must be filled to preserve genome stability. Using a DNA fiber approach to directly monitor gap filling, we studied the post-replicative mechanisms that fill the ssDNA gaps generated in cisplatin-treated cells upon increased PRIMPOL expression or when replication fork reversal is defective because of SMARCAL1 inactivation or PARP inhibition. We found that a mechanism dependent on the E3 ubiquitin ligase RAD18, PCNA monoubiquitination, and the REV1 and POLζ translesion synthesis polymerases promotes gap filling in G2. The E2-conjugating enzyme UBC13, the RAD51 recombinase, and REV1-POLζ are instead responsible for gap filling in S, suggesting that temporally distinct pathways of gap filling operate throughout the cell cycle. Furthermore, we found that BRCA1 and BRCA2 promote gap filling by limiting MRE11 activity and that simultaneously targeting fork reversal and gap filling enhances chemosensitivity in BRCA-deficient cells.
Keywords: BRCA1; BRCA2; DNA damage tolerance; DNA replication; PRIMPOL; genome stability; post-replicative repair; replication stress; ssDNA gaps; translesion synthesis polymerases.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Ashworth A, and Lord CJ (2018). Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol 15, 564–576. - PubMed
-
- Bailly V, Lamb J, Sung P, Prakash S, and Prakash L (1994). Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8, 811–820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

