Exercise as a model to identify microRNAs linked to human cognition: a role for microRNA-409 and microRNA-501
- PMID: 34625536
- PMCID: PMC8501071
- DOI: 10.1038/s41398-021-01627-w
Exercise as a model to identify microRNAs linked to human cognition: a role for microRNA-409 and microRNA-501
Abstract
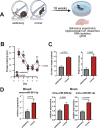
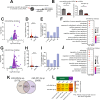
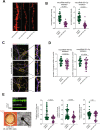
MicroRNAs have been linked to synaptic plasticity and memory function and are emerging as potential biomarkers and therapeutic targets for cognitive diseases. Most of these data stem from the analysis of model systems or postmortem tissue from patients which mainly represents an advanced stage of pathology. Due to the in-accessibility of human brain tissue upon experimental manipulation, it is still challenging to identify microRNAs relevant to human cognition, which is however a key step for future translational studies. Here, we employ exercise as an experimental model for memory enhancement in healthy humans with the aim to identify microRNAs linked to memory function. By analyzing the circulating smallRNAome we find a cluster of 18 microRNAs that are highly correlated to cognition. MicroRNA-409-5p and microRNA-501-3p were the most significantly regulated candidates. Functional analysis revealed that the two microRNAs are important for neuronal integrity, synaptic plasticity, and morphology. In conclusion, we provide a novel approach to identify microRNAs linked to human memory function.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

