Stage-resolved Hi-C analyses reveal meiotic chromosome organizational features influencing homolog alignment
- PMID: 34625553
- PMCID: PMC8501046
- DOI: 10.1038/s41467-021-26033-0
Stage-resolved Hi-C analyses reveal meiotic chromosome organizational features influencing homolog alignment
Abstract
During meiosis, chromosomes exhibit dramatic changes in morphology and intranuclear positioning. How these changes influence homolog pairing, alignment, and recombination remain elusive. Using Hi-C, we systematically mapped 3D genome architecture throughout all meiotic prophase substages during mouse spermatogenesis. Our data uncover two major chromosome organizational features varying along the chromosome axis during early meiotic prophase, when homolog alignment occurs. First, transcriptionally active and inactive genomic regions form alternating domains consisting of shorter and longer chromatin loops, respectively. Second, the force-transmitting LINC complex promotes the alignment of ends of different chromosomes over a range of up to 20% of chromosome length. Both features correlate with the pattern of homolog interactions and the distribution of recombination events. Collectively, our data reveal the influences of transcription and force on meiotic chromosome structure and suggest chromosome organization may provide an infrastructure for the modulation of meiotic recombination in higher eukaryotes.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


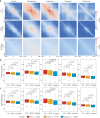
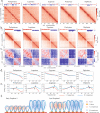
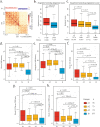
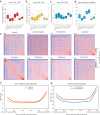
References
-
- Gerton, J. L. & Hawley, R. S. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 10.1038/nrg1614 (2005). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

