Fatty acids can inhibit Staphylococcus aureus SaeS activity at the membrane independent of alterations in respiration
- PMID: 34626146
- PMCID: PMC8898382
- DOI: 10.1111/mmi.14830
Fatty acids can inhibit Staphylococcus aureus SaeS activity at the membrane independent of alterations in respiration
Abstract
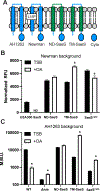
In Staphylococcus aureus, the two-component system SaeRS is responsible for regulating various virulence factors essential for the success of this pathogen. SaeRS can be stimulated by neutrophil-derived products but has also recently been shown to be inactivated by the presence of free fatty acids. A mechanism for how fatty acids negatively impacts SaeRS has not been described. We found that unsaturated fatty acids, as well as fatty acids not commonly found in Staphylococcal membranes, prevent the activation of SaeRS at a lower concentration than their saturated counterparts. These fatty acids can negatively impact SaeRS without altering the respiratory capacity of the bacterium. To uncover a potential mechanism for how fatty acids impact SaeRS function/activity, we utilized a naturally occurring point mutation found in S. aureus as well as chimeric SaeS proteins. Using these tools, we identified that the native transmembrane domains of SaeS dictate the transcriptional response to fatty acids in S. aureus. Our data support a model where free fatty acids alter the activity of the two-component system SaeRS directly through the sensor kinase SaeS and is dependent on the transmembrane domains of the protein.
Keywords: S. aureus; MRSA; SaeRS; two-component system; virulence factor.
© 2021 John Wiley & Sons Ltd.
Figures
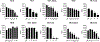
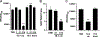
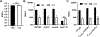

Similar articles
-
Treatment of Staphylococcus aureus with environmentally relevant concentrations of triclosan activates SaeRS-dependent virulence factor expression.Antimicrob Agents Chemother. 2025 Aug 6;69(8):e0172824. doi: 10.1128/aac.01728-24. Epub 2025 Jun 18. Antimicrob Agents Chemother. 2025. PMID: 40531055 Free PMC article.
-
Role of Fatty Acid Kinase in Cellular Lipid Homeostasis and SaeRS-Dependent Virulence Factor Expression in Staphylococcus aureus.mBio. 2017 Aug 1;8(4):e00988-17. doi: 10.1128/mBio.00988-17. mBio. 2017. PMID: 28765222 Free PMC article.
-
VfrB Is a Key Activator of the Staphylococcus aureus SaeRS Two-Component System.J Bacteriol. 2017 Feb 14;199(5):e00828-16. doi: 10.1128/JB.00828-16. Print 2017 Mar 1. J Bacteriol. 2017. PMID: 28031278 Free PMC article.
-
The SaeRS Two-Component System of Staphylococcus aureus.Genes (Basel). 2016 Oct 3;7(10):81. doi: 10.3390/genes7100081. Genes (Basel). 2016. PMID: 27706107 Free PMC article. Review.
-
Metabolic control of virulence factor production in Staphylococcus aureus.Curr Opin Microbiol. 2020 Jun;55:81-87. doi: 10.1016/j.mib.2020.03.004. Epub 2020 May 7. Curr Opin Microbiol. 2020. PMID: 32388086 Free PMC article. Review.
Cited by
-
Treatment of Staphylococcus aureus with environmentally relevant concentrations of triclosan activates SaeRS-dependent virulence factor expression.Antimicrob Agents Chemother. 2025 Aug 6;69(8):e0172824. doi: 10.1128/aac.01728-24. Epub 2025 Jun 18. Antimicrob Agents Chemother. 2025. PMID: 40531055 Free PMC article.
-
Two-Component Systems of S. aureus: Signaling and Sensing Mechanisms.Genes (Basel). 2021 Dec 23;13(1):34. doi: 10.3390/genes13010034. Genes (Basel). 2021. PMID: 35052374 Free PMC article. Review.
-
Pyrimidine sufficiency is required for Sae two-component system signaling in Staphylococcus aureus.J Bacteriol. 2025 Aug 21;207(8):e0011525. doi: 10.1128/jb.00115-25. Epub 2025 Jul 21. J Bacteriol. 2025. PMID: 40689635 Free PMC article.
-
Regulation of Bacterial Two-Component Systems by Cardiolipin.Infect Immun. 2023 Apr 18;91(4):e0004623. doi: 10.1128/iai.00046-23. Epub 2023 Mar 28. Infect Immun. 2023. PMID: 36975788 Free PMC article.
-
Regulation of the Sae Two-Component System by Branched-Chain Fatty Acids in Staphylococcus aureus.mBio. 2022 Oct 26;13(5):e0147222. doi: 10.1128/mbio.01472-22. Epub 2022 Sep 22. mBio. 2022. PMID: 36135382 Free PMC article.
References
-
- Adhikari RP, and Novick RP (2008) Regulatory organization of the staphylococcal sae locus. Microbiology 154: 949–959. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

