The human hepatocyte TXG-MAPr: gene co-expression network modules to support mechanism-based risk assessment
- PMID: 34626214
- PMCID: PMC8536636
- DOI: 10.1007/s00204-021-03141-w
The human hepatocyte TXG-MAPr: gene co-expression network modules to support mechanism-based risk assessment
Abstract
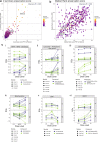
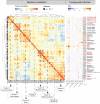
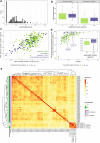
Mechanism-based risk assessment is urged to advance and fully permeate into current safety assessment practices, possibly at early phases of drug safety testing. Toxicogenomics is a promising source of mechanisms-revealing data, but interpretative analysis tools specific for the testing systems (e.g. hepatocytes) are lacking. In this study, we present the TXG-MAPr webtool (available at https://txg-mapr.eu/WGCNA_PHH/TGGATEs_PHH/ ), an R-Shiny-based implementation of weighted gene co-expression network analysis (WGCNA) obtained from the Primary Human Hepatocytes (PHH) TG-GATEs dataset. The 398 gene co-expression networks (modules) were annotated with functional information (pathway enrichment, transcription factor) to reveal their mechanistic interpretation. Several well-known stress response pathways were captured in the modules, were perturbed by specific stressors and showed preservation in rat systems (rat primary hepatocytes and rat in vivo liver), with the exception of DNA damage and oxidative stress responses. A subset of 87 well-annotated and preserved modules was used to evaluate mechanisms of toxicity of endoplasmic reticulum (ER) stress and oxidative stress inducers, including cyclosporine A, tunicamycin and acetaminophen. In addition, module responses can be calculated from external datasets obtained with different hepatocyte cells and platforms, including targeted RNA-seq data, therefore, imputing biological responses from a limited gene set. As another application, donors' sensitivity towards tunicamycin was investigated with the TXG-MAPr, identifying higher basal level of intrinsic immune response in donors with pre-existing liver pathology. In conclusion, we demonstrated that gene co-expression analysis coupled to an interactive visualization environment, the TXG-MAPr, is a promising approach to achieve mechanistic relevant, cross-species and cross-platform evaluation of toxicogenomic data.
Keywords: DILI; ER stress; Mechanism-based risk assessment; PHH; WGCNA.
© 2021. The Author(s).
Conflict of interest statement
JSR received funding from GSK and Sanofi and consultant fees from Travere Therapeutics.
Figures



Similar articles
-
Characterisation of the NRF2 transcriptional network and its response to chemical insult in primary human hepatocytes: implications for prediction of drug-induced liver injury.Arch Toxicol. 2019 Feb;93(2):385-399. doi: 10.1007/s00204-018-2354-1. Epub 2018 Nov 13. Arch Toxicol. 2019. PMID: 30426165 Free PMC article.
-
Utilization of causal reasoning of hepatic gene expression in rats to identify molecular pathways of idiosyncratic drug-induced liver injury.Toxicol Sci. 2014 Jan;137(1):234-48. doi: 10.1093/toxsci/kft232. Epub 2013 Oct 17. Toxicol Sci. 2014. PMID: 24136188
-
Toxicogenomic module associations with pathogenesis: a network-based approach to understanding drug toxicity.Pharmacogenomics J. 2018 May 22;18(3):377-390. doi: 10.1038/tpj.2017.17. Epub 2017 Apr 25. Pharmacogenomics J. 2018. PMID: 28440344
-
Application of toxicogenomics in hepatic systems toxicology for risk assessment: acetaminophen as a case study.Toxicol Appl Pharmacol. 2011 Jan 15;250(2):96-107. doi: 10.1016/j.taap.2010.10.013. Epub 2010 Oct 21. Toxicol Appl Pharmacol. 2011. PMID: 20970440 Review.
-
The application of omics-based human liver platforms for investigating the mechanism of drug-induced hepatotoxicity in vitro.Arch Toxicol. 2019 Nov;93(11):3067-3098. doi: 10.1007/s00204-019-02585-5. Epub 2019 Oct 5. Arch Toxicol. 2019. PMID: 31586243 Review.
Cited by
-
Analyses of Transcriptomics Cell Signalling for Pre-Screening Applications in the Integrated Approach for Testing and Assessment of Non-Genotoxic Carcinogens.Int J Mol Sci. 2022 Oct 22;23(21):12718. doi: 10.3390/ijms232112718. Int J Mol Sci. 2022. PMID: 36361516 Free PMC article. Review.
-
Searching for LINCS to Stress: Using Text Mining to Automate Reference Chemical Curation.Chem Res Toxicol. 2024 Jun 17;37(6):878-893. doi: 10.1021/acs.chemrestox.3c00335. Epub 2024 May 13. Chem Res Toxicol. 2024. PMID: 38736322 Free PMC article.
-
A network-based transcriptomic landscape of HepG2 cells uncovering causal gene-cytotoxicity interactions underlying drug-induced liver injury.Toxicol Sci. 2024 Feb 28;198(1):14-30. doi: 10.1093/toxsci/kfad121. Toxicol Sci. 2024. PMID: 38015832 Free PMC article.
-
Assessment of pulmonary fibrosis using weighted gene co-expression network analysis.Front Toxicol. 2024 Oct 24;6:1465704. doi: 10.3389/ftox.2024.1465704. eCollection 2024. Front Toxicol. 2024. PMID: 39512679 Free PMC article.
-
Assessment of Drug-Induced Liver Injury through Cell Morphology and Gene Expression Analysis.Chem Res Toxicol. 2023 Sep 18;36(9):1456-1470. doi: 10.1021/acs.chemrestox.2c00381. Epub 2023 Aug 31. Chem Res Toxicol. 2023. PMID: 37652439 Free PMC article.
References
-
- Alexa A, Rahnenführer J (2007) topGO: Enrichment Analysis for Gene Ontology. R package
-
- Björnsson ES. Global epidemiology of drug-induced liver injury (DILI) Curr Hepatol Rep. 2019;18(3):274–279. doi: 10.1007/s11901-019-00475-z. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

