A new SARS-CoV-2 variant with high lethality poorly detected by RT-PCR on nasopharyngeal samples: an observational study
- PMID: 34627988
- PMCID: PMC8496927
- DOI: 10.1016/j.cmi.2021.09.035
A new SARS-CoV-2 variant with high lethality poorly detected by RT-PCR on nasopharyngeal samples: an observational study
Abstract
Objectives: In early January 2021 an outbreak of nosocomial cases of coronavirus disease 2019 (COVID-19) emerged in Western France; RT-PCR tests were repeatedly negative on nasopharyngeal samples but positive on lower respiratory tract samples. Whole-genome sequencing (WGS) revealed a new variant, currently defining a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.616. In March, the WHO classified this as a 'variant under investigation' (VUI). We analysed the characteristics and outcomes of COVID-19 cases related to this new variant.
Methods: Clinical, virological, and radiological data were retrospectively collected from medical charts in the two hospitals involved. We enrolled those inpatients with: (a) positive SARS-CoV-2 RT-PCR on a respiratory sample, (b) seroconversion with anti-SARS-CoV-2 IgG/IgM, or (c) suggestive symptoms and typical features of COVID-19 on a chest CT scan. Cases were categorized as B.1.616, a variant of concern (VOC), or unknown.
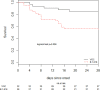
Results: From 1st January to 24th March 2021, 114 patients fulfilled the inclusion criteria: B.1.616 (n = 39), VOC (n = 32), and unknown (n = 43). B.1.616-related cases were older than VOC-related cases (81 years, interquartile range (IQR) 73-88 versus 73 years, IQR 67-82, p < 0.05) and their first RT-PCR tests were rarely positive (6/39, 15% versus 31/32, 97%, p < 0.05). The B.1.616 variant was independently associated with severe disease (multivariable Cox model HR 4.0, 95%CI 1.5-10.9) and increased lethality (28-day mortality 18/39 (46%) for B.1.616 versus 5/32 (16%) for VOC, p = 0.006).
Conclusion: We report a nosocomial outbreak of COVID-19 cases related to a new variant, B.1.616, which is poorly detected by RT-PCR on nasopharyngeal samples and is associated with high lethality.
Keywords: COVID-19; Coronavirus infections/epidemiology; Coronavirus infections/virology; SARS-CoV-2 variants; Severity of illness index.
Copyright © 2021 European Society of Clinical Microbiology and Infectious Diseases. Published by Elsevier Ltd. All rights reserved.
Figures


References
-
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. - PubMed
-
- Weekly epidemiological update on COVID-19—27 April 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on... n.d.
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

