Inter-species interactions alter antibiotic efficacy in bacterial communities
- PMID: 34628478
- PMCID: PMC8857223
- DOI: 10.1038/s41396-021-01130-6
Inter-species interactions alter antibiotic efficacy in bacterial communities
Abstract
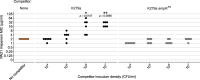
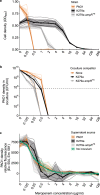
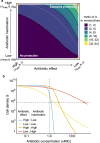
The efficacy of antibiotic treatments targeting polymicrobial communities is not well predicted by conventional in vitro susceptibility testing based on determining minimum inhibitory concentration (MIC) in monocultures. One reason for this is that inter-species interactions can alter the community members' susceptibility to antibiotics. Here we quantify, and identify mechanisms for, community-modulated changes of efficacy for clinically relevant antibiotics against the pathogen Pseudomonas aeruginosa in model cystic fibrosis (CF) lung communities derived from clinical samples. We demonstrate that multi-drug resistant Stenotrophomonas maltophilia can provide high levels of antibiotic protection to otherwise sensitive P. aeruginosa. Exposure protection to imipenem was provided by chromosomally encoded metallo-β-lactamase that detoxified the environment; protection was dependent upon S. maltophilia cell density and was provided by S. maltophilia strains isolated from CF sputum, increasing the MIC of P. aeruginosa by up to 16-fold. In contrast, the presence of S. maltophilia provided no protection against meropenem, another routinely used carbapenem. Mathematical ordinary differential equation modelling shows that the level of exposure protection provided against different carbapenems can be explained by differences in antibiotic efficacy and inactivation rate. Together, these findings reveal that exploitation of pre-occurring antimicrobial resistance, and inter-specific competition, can have large impacts on pathogen antibiotic susceptibility, highlighting the importance of microbial ecology for designing successful antibiotic treatments for multispecies communities.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Finding protection in the community.Nat Rev Microbiol. 2022 Jan;20(1):2. doi: 10.1038/s41579-021-00654-0. Nat Rev Microbiol. 2022. PMID: 34697495 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

