M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway
- PMID: 34628486
- PMCID: PMC9984538
- DOI: 10.1038/s41418-021-00888-8
M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway
Erratum in
-
Correction to: M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway.Cell Death Differ. 2023 Mar;30(3):856. doi: 10.1038/s41418-022-01075-z. Cell Death Differ. 2023. PMID: 36216972 Free PMC article. No abstract available.
Abstract
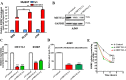
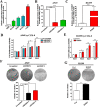
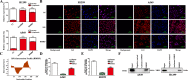
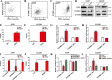
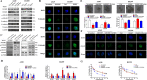
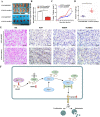
Non-small cell lung cancer (NSCLC) has the highest mortality rate among all malignancies worldwide. The role of long noncoding RNAs (lncRNAs) in the progression of cancers is a contemporary research hotspot. Based on an integrative analysis of The Cancer Genome Atlas database, we identified lncRNA-RNA Component of Mitochondrial RNA Processing Endoribonuclease (RMRP) as one of the most highly upregulated lncRNAs that are associated with poor survival in NSCLC. Furthermore, N(6)-methyladenosine (m6A) was highly enriched within RMRP and enhanced its RNA stability. In vitro and in vivo experiments showed that RMRP promoted NSCLC cell proliferation, invasion, and migration. In terms of mechanism, RMRP recruited YBX1 to the TGFBR1 promotor region, leading to upregulation of the transcription of TGFBR1. The TGFBR1/SMAD2/SMAD3 pathway was also regulated by RMRP. In addition, RMRP promoted the cancer stem cells properties and epithelial mesenchymal transition, which promote the resistance to radiation therapy and cisplatin. Clinical data further confirmed a positive correlation between RMRP and TGFBR1. In short, our work reveals that m6A RNA methylation-mediated RMRP stability renders proliferation and progression of NSCLC through regulating TGFBR1/SMAD2/SMAD3 pathway.
© 2021. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

