Chip Protein U-Box Domain Truncation Affects Purkinje Neuron Morphology and Leads to Behavioral Changes in Zebrafish
- PMID: 34630034
- PMCID: PMC8497888
- DOI: 10.3389/fnmol.2021.723912
Chip Protein U-Box Domain Truncation Affects Purkinje Neuron Morphology and Leads to Behavioral Changes in Zebrafish
Abstract
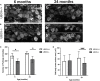
The ubiquitin ligase CHIP (C-terminus of Hsc70-interacting protein) is encoded by STUB1 and promotes ubiquitination of misfolded and damaged proteins. CHIP deficiency has been linked to several diseases, and mutations in the human STUB1 gene are associated with recessive and dominant forms of spinocerebellar ataxias (SCAR16/SCA48). Here, we examine the effects of impaired CHIP ubiquitin ligase activity in zebrafish (Danio rerio). We characterized the zebrafish stub1 gene and Chip protein, and generated and characterized a zebrafish mutant causing truncation of the Chip functional U-box domain. Zebrafish stub1 has a high degree of conservation with mammalian orthologs and was detected in a wide range of tissues in adult stages, with highest expression in brain, eggs, and testes. In the brain, stub1 mRNA was predominantly detected in the cerebellum, including the Purkinje cell layer and granular layer. Recombinant wild-type zebrafish Chip showed ubiquitin ligase activity highly comparable to human CHIP, while the mutant Chip protein showed impaired ubiquitination of the Hsc70 substrate and Chip itself. In contrast to SCAR16/SCA48 patients, no gross cerebellar atrophy was evident in mutant fish, however, these fish displayed reduced numbers and sizes of Purkinje cell bodies and abnormal organization of Purkinje cell dendrites. Mutant fish also had decreased total 26S proteasome activity in the brain and showed behavioral changes. In conclusion, truncation of the Chip U-box domain leads to impaired ubiquitin ligase activity and behavioral and anatomical changes in zebrafish, illustrating the potential of zebrafish to study STUB1-mediated diseases.
Keywords: CHIP; Purkinje cells; SCAR16; STUB1; anxiety-like behavior; zebrafish.
Copyright © 2021 Pakdaman, Denker, Austad, Norton, Rolfsnes, Bindoff, Tzoulis, Aukrust, Knappskog, Johansson and Ellingsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Genetic Dominant Variants in STUB1, Segregating in Families with SCA48, Display In Vitro Functional Impairments Indistinctive from Recessive Variants Associated with SCAR16.Int J Mol Sci. 2021 May 30;22(11):5870. doi: 10.3390/ijms22115870. Int J Mol Sci. 2021. PMID: 34070858 Free PMC article.
-
Clinical and functional characterization of a novel STUB1 mutation in a Chinese spinocerebellar ataxia 48 pedigree.Orphanet J Rare Dis. 2024 Dec 20;19(1):471. doi: 10.1186/s13023-024-03456-8. Orphanet J Rare Dis. 2024. PMID: 39707479 Free PMC article.
-
Clinical and functional characterization of a novel STUB1 frameshift mutation in autosomal dominant spinocerebellar ataxia type 48 (SCA48).J Biomed Sci. 2021 Sep 26;28(1):65. doi: 10.1186/s12929-021-00763-1. J Biomed Sci. 2021. PMID: 34565360 Free PMC article.
-
Spinocerebellar ataxia type 48: last but not least.Neurol Sci. 2020 Sep;41(9):2423-2432. doi: 10.1007/s10072-020-04408-3. Epub 2020 Apr 27. Neurol Sci. 2020. PMID: 32342324 Review.
-
STUB1/CHIP: New insights in cancer and immunity.Biomed Pharmacother. 2023 Sep;165:115190. doi: 10.1016/j.biopha.2023.115190. Epub 2023 Jul 26. Biomed Pharmacother. 2023. PMID: 37506582 Review.
Cited by
-
CHIP and aging: a key regulator of proteostasis and cellular senescence.Biogerontology. 2025 May 5;26(3):104. doi: 10.1007/s10522-025-10247-6. Biogerontology. 2025. PMID: 40323531 Review.
References
-
- Anderson L. G., Meeker R. B., Poulton W. E., Huang D. Y. (2010). Brain distribution of carboxy terminus of Hsc70-interacting protein (CHIP) and its nuclear translocation in cultured cortical neurons following heat stress or oxygen-glucose deprivation. Cell Stress Chaperones 15 487–495. 10.1007/s12192-009-0162-5 - DOI - PMC - PubMed
-
- Bettencourt C., de Yébenes J. G., ópez-Sendón J. L. L., Shomroni O., Zhang X., Qian S. B., I, et al. (2015). Clinical and neuropathological features of spastic ataxia in a spanish family with novel compound heterozygous mutations in STUB1. Cerebellum 14 378–381. 10.1007/s12311-014-0643-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

