Novel Functional Properties of Missense Mutations in the Glycine Receptor β Subunit in Startle Disease
- PMID: 34630038
- PMCID: PMC8498107
- DOI: 10.3389/fnmol.2021.745275
Novel Functional Properties of Missense Mutations in the Glycine Receptor β Subunit in Startle Disease
Abstract
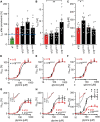
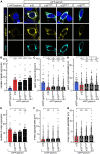
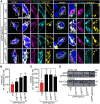
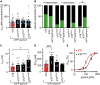
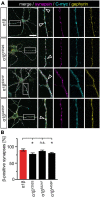
Startle disease is a rare disorder associated with mutations in GLRA1 and GLRB, encoding glycine receptor (GlyR) α1 and β subunits, which enable fast synaptic inhibitory transmission in the spinal cord and brainstem. The GlyR β subunit is important for synaptic localization via interactions with gephyrin and contributes to agonist binding and ion channel conductance. Here, we have studied three GLRB missense mutations, Y252S, S321F, and A455P, identified in startle disease patients. For Y252S in M1 a disrupted stacking interaction with surrounding aromatic residues in M3 and M4 is suggested which is accompanied by an increased EC50 value. By contrast, S321F in M3 might stabilize stacking interactions with aromatic residues in M1 and M4. No significant differences in glycine potency or efficacy were observed for S321F. The A455P variant was not predicted to impact on subunit folding but surprisingly displayed increased maximal currents which were not accompanied by enhanced surface expression, suggesting that A455P is a gain-of-function mutation. All three GlyR β variants are trafficked effectively with the α1 subunit through intracellular compartments and inserted into the cellular membrane. In vivo, the GlyR β subunit is transported together with α1 and the scaffolding protein gephyrin to synaptic sites. The interaction of these proteins was studied using eGFP-gephyrin, forming cytosolic aggregates in non-neuronal cells. eGFP-gephyrin and β subunit co-expression resulted in the recruitment of both wild-type and mutant GlyR β subunits to gephyrin aggregates. However, a significantly lower number of GlyR β aggregates was observed for Y252S, while for mutants S321F and A455P, the area and the perimeter of GlyR β subunit aggregates was increased in comparison to wild-type β. Transfection of hippocampal neurons confirmed differences in GlyR-gephyrin clustering with Y252S and A455P, leading to a significant reduction in GlyR β-positive synapses. Although none of the mutations studied is directly located within the gephyrin-binding motif in the GlyR β M3-M4 loop, we suggest that structural changes within the GlyR β subunit result in differences in GlyR β-gephyrin interactions. Hence, we conclude that loss- or gain-of-function, or alterations in synaptic GlyR clustering may underlie disease pathology in startle disease patients carrying GLRB mutations.
Keywords: GLRB; gephyrin; glycine receptor; hyperekplexia; startle disease.
Copyright © 2021 Piro, Eckes, Kasaragod, Sommer, Harvey, Schaefer and Villmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Isoform heterogeneity of the human gephyrin gene (GPHN), binding domains to the glycine receptor, and mutation analysis in hyperekplexia.J Biol Chem. 2003 Jul 4;278(27):24688-96. doi: 10.1074/jbc.M301070200. Epub 2003 Apr 8. J Biol Chem. 2003. PMID: 12684523
-
A Missense Mutation A384P Associated with Human Hyperekplexia Reveals a Desensitization Site of Glycine Receptors.J Neurosci. 2018 Mar 14;38(11):2818-2831. doi: 10.1523/JNEUROSCI.0674-16.2018. Epub 2018 Feb 13. J Neurosci. 2018. PMID: 29440552 Free PMC article.
-
Disruption of a Structurally Important Extracellular Element in the Glycine Receptor Leads to Decreased Synaptic Integration and Signaling Resulting in Severe Startle Disease.J Neurosci. 2017 Aug 16;37(33):7948-7961. doi: 10.1523/JNEUROSCI.0009-17.2017. Epub 2017 Jul 19. J Neurosci. 2017. PMID: 28724750 Free PMC article.
-
Impaired Glycine Receptor Trafficking in Neurological Diseases.Front Mol Neurosci. 2018 Aug 21;11:291. doi: 10.3389/fnmol.2018.00291. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30186111 Free PMC article. Review.
-
Milestone Review: Unlocking the Proteomics of Glycine Receptor Complexes.J Neurochem. 2025 Apr;169(4):e70061. doi: 10.1111/jnc.70061. J Neurochem. 2025. PMID: 40285371 Free PMC article. Review.
Cited by
-
Glycine Receptor β-Targeting Autoantibodies Contribute to the Pathology of Autoimmune Diseases.Neurol Neuroimmunol Neuroinflamm. 2024 Mar;11(2):e200187. doi: 10.1212/NXI.0000000000200187. Epub 2024 Jan 12. Neurol Neuroimmunol Neuroinflamm. 2024. PMID: 38215349 Free PMC article.
-
Role of the Glycine Receptor β Subunit in Synaptic Localization and Pathogenicity in Severe Startle Disease.J Neurosci. 2024 Jan 10;44(2):e0837232023. doi: 10.1523/JNEUROSCI.0837-23.2023. J Neurosci. 2024. PMID: 37963764 Free PMC article.
-
Modulatory Actions of the Glycine Receptor β Subunit on the Positive Allosteric Modulation of Ethanol in α2 Containing Receptors.Front Mol Neurosci. 2021 Nov 18;14:763868. doi: 10.3389/fnmol.2021.763868. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34867189 Free PMC article.
-
Loss, Gain and Altered Function of GlyR α2 Subunit Mutations in Neurodevelopmental Disorders.Front Mol Neurosci. 2022 Apr 29;15:886729. doi: 10.3389/fnmol.2022.886729. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35571374 Free PMC article.
-
Startle Disease: New Molecular Insights into an Old Neurological Disorder.Neuroscientist. 2023 Dec;29(6):767-781. doi: 10.1177/10738584221104724. Epub 2022 Jun 25. Neuroscientist. 2023. PMID: 35754344 Free PMC article. Review.
References
-
- Aboheimed G., Alrasheed M., Salih M., Alowain A., Alsagob M., Almass R., et al. (2019). Novel glycine receptor variants identified in startle disease patients. Eur. J. Hum. Genet. 27 1466–1467. - PubMed
-
- Atak S., Langlhofer G., Schaefer N., Kessler D., Meiselbach H., Delto C., et al. (2015). Disturbances of ligand potency and enhanced degradation of the human glycine receptor at affected positions G160 and T162 originally identified in patients suffering from hyperekplexia. Front. Mol. Neurosci. 8:79. 10.3389/fnmol.2015.00079 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

