Bifidobacterium Strains Present Distinct Effects on the Control of Alveolar Bone Loss in a Periodontitis Experimental Model
- PMID: 34630089
- PMCID: PMC8497694
- DOI: 10.3389/fphar.2021.713595
Bifidobacterium Strains Present Distinct Effects on the Control of Alveolar Bone Loss in a Periodontitis Experimental Model
Abstract
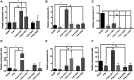
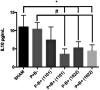
Periodontitis is an inflammatory disease induced by a dysbiotic oral microbiome. Probiotics of the genus Bifidobacterium may restore the symbiotic microbiome and modulate the immune response, leading to periodontitis control. We evaluated the effect of two strains of Bifidobacterium able to inhibit Porphyromonas gingivalis interaction with host cells and biofilm formation, but with distinct immunomodulatory properties, in a mice periodontitis model. Experimental periodontitis (P+) was induced in C57Bl/6 mice by a microbial consortium of human oral organisms. B. bifidum 1622A [B+ (1622)] and B. breve 1101A [B+ (1101)] were orally inoculated for 45 days. Alveolar bone loss and inflammatory response in gingival tissues were determined. The microbial consortium induced alveolar bone loss in positive control (P + B-), as demonstrated by microtomography analysis, although P. gingivalis was undetected in oral biofilms at the end of the experimental period. TNF-α and IL-10 serum levels, and Treg and Th17 populations in gingiva of SHAM and P + B- groups did not differ. B. bifidum 1622A, but not B. breve 1101A, controlled bone destruction in P+ mice. B. breve 1101A upregulated transcription of Il-1β, Tnf-α, Tlr2, Tlr4, and Nlrp3 in P-B+(1101), which was attenuated by the microbial consortium [P + B+(1101)]. All treatments downregulated transcription of Il-17, although treatment with B. breve 1101A did not yield such low levels of transcripts as seen for the other groups. B. breve 1101A increased Th17 population in gingival tissues [P-B+ (1101) and P + B+ (1101)] compared to SHAM and P + B-. Administration of both bifidobacteria resulted in serum IL-10 decreased levels. Our data indicated that the beneficial effect of Bifidobacterium is not a common trait of this genus, since B. breve 1101A induced an inflammatory profile in gingival tissues and did not prevent alveolar bone loss. However, the properties of B. bifidum 1622A suggest its potential to control periodontitis.
Keywords: Bifidobacterium; P. gingivalis; immune modulation; periodontitis; probiotics.
Copyright © 2021 Shimabukuro, Cataruci, Ishikawa, Oliveira, Kawamoto, Ando-Suguimoto, Albuquerque-Souza, Nicoli, Ferreira, de Lima, Bueno, da Silva, Silva, Messora, Camara, Simionato and Mayer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Albuquerque-Souza E., Balzarini D., Ando-Suguimoto E. S., Ishikawa K. H., Simionato M. R. L., Holzhausen M., et al. (2019). Probiotics Alter the Immune Response of Gingival Epithelial Cells Challenged by Porphyromonas gingivalis . J. Periodontal Res. 54 (2), 115–127. 10.1111/jre.12608 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

