Live-cell imaging of microRNA expression with post-transcriptional feedback control
- PMID: 34631284
- PMCID: PMC8479275
- DOI: 10.1016/j.omtn.2021.08.018
Live-cell imaging of microRNA expression with post-transcriptional feedback control
Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that regulate complex gene expression networks in eukaryotic cells. Because of their unique expression patterns, miRNAs are potential molecular markers for specific cell states. Although a system capable of imaging miRNA in living cells is needed to visually detect miRNA expression, very few fluorescence signal-on sensors that respond to expression of target miRNA (miR-ON sensors) are available. Here we report an miR-ON sensor containing a bidirectional promoter-driven Csy4 endoribonuclease and green fluorescent protein, ZsGreen1, for live-cell imaging of miRNAs with post-transcriptional feedback control. Csy4-assisted miR-ON (Csy4-miR-ON) sensors generate negligible background but respond sensitively to target miRNAs, allowing high-contrast fluorescence detection of miRNAs in various human cells. We show that Csy4-miR-ON sensors enabled imaging of various miRNAs, including miR-21, miR-302a, and miR-133, in vitro as well as in vivo. This robust tool can be used to evaluate miRNA expression in diverse biological and medical applications.
Keywords: Csy4; fluorescent protein; live-cell imaging; microRNA; post-transcriptional feedback control.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


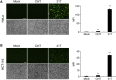
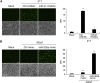
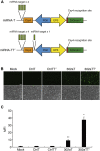

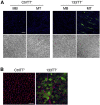
Similar articles
-
Time-lapse imaging of microRNA activity reveals the kinetics of microRNA activation in single living cells.Sci Rep. 2017 Oct 3;7(1):12642. doi: 10.1038/s41598-017-12879-2. Sci Rep. 2017. PMID: 28974737 Free PMC article.
-
Early-onset preeclampsia, plasma microRNAs, and endothelial cell function.Am J Obstet Gynecol. 2020 May;222(5):497.e1-497.e12. doi: 10.1016/j.ajog.2019.11.1286. Epub 2019 Dec 10. Am J Obstet Gynecol. 2020. PMID: 31836544
-
Optimization of a microRNA expression vector for function analysis of microRNA.J Control Release. 2011 Feb 28;150(1):94-101. doi: 10.1016/j.jconrel.2010.12.001. Epub 2010 Dec 10. J Control Release. 2011. PMID: 21146569
-
Application of the microRNA-302/367 cluster in cancer therapy.Cancer Sci. 2020 Apr;111(4):1065-1075. doi: 10.1111/cas.14317. Epub 2020 Feb 13. Cancer Sci. 2020. PMID: 31957939 Free PMC article. Review.
-
The Role and Function of microRNA in the Pathogenesis of Multiple Myeloma.Cancers (Basel). 2019 Nov 6;11(11):1738. doi: 10.3390/cancers11111738. Cancers (Basel). 2019. PMID: 31698726 Free PMC article. Review.
Cited by
-
Live-cell RNA imaging with the inactivated endonuclease Csy4 enables new insights into plant virus transport through plasmodesmata.PLoS Pathog. 2025 Apr 9;21(4):e1013049. doi: 10.1371/journal.ppat.1013049. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40203052 Free PMC article.
-
Sensing and guiding cell-state transitions by using genetically encoded endoribonuclease-mediated microRNA sensors.Nat Biomed Eng. 2024 Dec;8(12):1730-1743. doi: 10.1038/s41551-024-01229-z. Epub 2024 Jul 9. Nat Biomed Eng. 2024. PMID: 38982158
-
An engineered ligand-responsive Csy4 endoribonuclease controls transgene expression from Sendai virus vectors.J Biol Eng. 2024 Jan 16;18(1):9. doi: 10.1186/s13036-024-00404-9. J Biol Eng. 2024. PMID: 38229076 Free PMC article.
-
Boolean modeling of mechanosensitive epithelial to mesenchymal transition and its reversal.iScience. 2023 Mar 2;26(4):106321. doi: 10.1016/j.isci.2023.106321. eCollection 2023 Apr 21. iScience. 2023. PMID: 36968076 Free PMC article.
-
A trigger-inducible split-Csy4 architecture for programmable RNA modulation.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1319. doi: 10.1093/nar/gkae1319. Nucleic Acids Res. 2025. PMID: 39817512 Free PMC article.
References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

