Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance
- PMID: 34631286
- PMCID: PMC8479277
- DOI: 10.1016/j.omtn.2021.08.020
Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance
Abstract
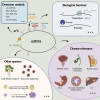
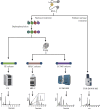
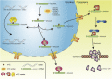
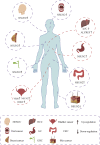
5-methylcytosine (m5C) post-transcriptional modifications affect the maturation, stability, and translation of the mRNA molecule. These modifications play an important role in many physiological and pathological processes, including stress response, tumorigenesis, tumor cell migration, embryogenesis, and viral replication. Recently, there has been a better understanding of the biological implications of m5C modification owing to the rapid development and optimization of detection technologies, including liquid chromatography-tandem mass spectrometry (LC-MS/MS) and RNA-BisSeq. Further, predictive models (such as PEA-m5C, m5C-PseDNC, and DeepMRMP) for the identification of potential m5C modification sites have also emerged. In this review, we summarize the current experimental detection methods and predictive models for mRNA m5C modifications, focusing on their advantages and limitations. We systematically surveyed the latest research on the effectors related to mRNA m5C modifications and their biological functions in multiple species. Finally, we discuss the physiological effects and pathological significance of m5C modifications in multiple diseases, as well as their therapeutic potential, thereby providing new perspectives for disease treatment and prognosis.
Keywords: 5-methylcytosine; RNA epigenetics; disease; mRNA; post-transcriptional modification; prediction.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

