Ambisense polarity of genome RNA of orthomyxoviruses and coronaviruses
- PMID: 34631475
- PMCID: PMC8474974
- DOI: 10.5501/wjv.v10.i5.256
Ambisense polarity of genome RNA of orthomyxoviruses and coronaviruses
Abstract
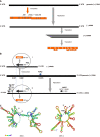
Influenza viruses and coronaviruses have linear single-stranded RNA genomes with negative and positive sense polarities and genes encoded in viral genomes are expressed in these viruses as positive and negative genes, respectively. Here we consider a novel gene identified in viral genomes in opposite direction, as positive in influenza and negative in coronaviruses, suggesting an ambisense genome strategy for both virus families. Noteworthy, the identified novel genes colocolized in the same RNA regions of viral genomes, where the previously known opposite genes are encoded, a so-called ambisense stacking architecture of genes in virus genome. It seems likely, that ambisense gene stacking in influenza and coronavirus families significantly increases genetic potential and virus diversity to extend virus-host adaptation pathways in nature. These data imply that ambisense viruses may have a multivirion mechanism, like "a dark side of the Moon", allowing production of the heterogeneous population of virions expressed through positive and negative sense genome strategies.
Keywords: Ambisense RNA; Coronavirus; Influenza; Virus diversity; Virus genes; Virus genome.
©The Author(s) 2021. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The author declares that he does do not have any conflict of interest. The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
The Unique Genome of the Virus and Alternative Strategies for its Realization.Acta Naturae. 2023 Apr-Jun;15(2):14-19. doi: 10.32607/actanaturae.11904. Acta Naturae. 2023. PMID: 37538802 Free PMC article.
-
Novel Negative Sense Genes in the RNA Genome of Coronaviruses.Dokl Biochem Biophys. 2021 May;496(1):27-31. doi: 10.1134/S1607672921010130. Epub 2021 Mar 10. Dokl Biochem Biophys. 2021. PMID: 33689070 Free PMC article.
-
Unique Bipolar Gene Architecture in the RNA Genome of Influenza A Virus.Biochemistry (Mosc). 2020 Mar;85(3):387-392. doi: 10.1134/S0006297920030141. Biochemistry (Mosc). 2020. PMID: 32564743 Free PMC article.
-
Expression strategies of ambisense viruses.Virus Res. 2003 Jun;93(2):141-50. doi: 10.1016/s0168-1702(03)00094-7. Virus Res. 2003. PMID: 12782362 Review.
-
Ambisense RNA viruses: positive and negative polarities combined in RNA virus genomes.Microbiol Sci. 1986 Jun;3(6):183-7. Microbiol Sci. 1986. PMID: 3153160 Review.
Cited by
-
The Unique Genome of the Virus and Alternative Strategies for its Realization.Acta Naturae. 2023 Apr-Jun;15(2):14-19. doi: 10.32607/actanaturae.11904. Acta Naturae. 2023. PMID: 37538802 Free PMC article.
-
RNA Viruses Linked to Eukaryotic Hosts in Thawed Permafrost.mSystems. 2022 Dec 20;7(6):e0058222. doi: 10.1128/msystems.00582-22. Epub 2022 Dec 1. mSystems. 2022. PMID: 36453933 Free PMC article.
References
-
- Zhirnov OP, Poyarkov SV, Vorob'eva IV, Safonova OA, Malyshev NA, Klenk HD. Segment NS of influenza A virus contains an additional gene NSP in positive-sense orientation. Dokl Biochem Biophys. 2007;414:127–133. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

