Recent Advances and Perspectives on Expanding the Chemical Diversity of Lasso Peptides
- PMID: 34631682
- PMCID: PMC8498205
- DOI: 10.3389/fbioe.2021.741364
Recent Advances and Perspectives on Expanding the Chemical Diversity of Lasso Peptides
Abstract
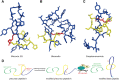
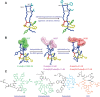
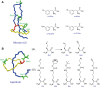
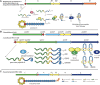
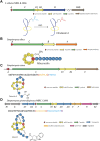
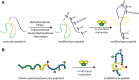
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a growing family of natural products that exhibit a range of structures and bioactivities. Initially assembled from the twenty proteinogenic amino acids in a ribosome-dependent manner, RiPPs assume their peculiar bioactive structures through various post-translational modifications. The essential modifications representative of each subfamily of RiPP are performed on a precursor peptide by the so-called processing enzymes; however, various tailoring enzymes can also embellish the precursor peptide or processed peptide with additional functional groups. Lasso peptides are an interesting subfamily of RiPPs characterized by their unique lariat knot-like structure, wherein the C-terminal tail is inserted through a macrolactam ring fused by an isopeptide bond between the N-terminal amino group and an acidic side chain. Until recently, relatively few lasso peptides were found to be tailored with extra functional groups. Nevertheless, the development of new routes to diversify lasso peptides and thus introduce novel or enhanced biological, medicinally relevant, or catalytic properties is appealing. In this review, we highlight several strategies through which lasso peptides have been successfully modified and provide a brief overview of the latest findings on the tailoring of these peptides. We also propose future directions for lasso peptide tailoring as well as potential applications for these peptides in hybrid catalyst design.
Keywords: biosynthesis; lasso peptide; natural products; post-translational modification; synthetic biology; tailoring enzymes.
Copyright © 2021 Wang, Fage, He, Mi, Yang, Li, An, Fan, Song, Zhu and Tong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

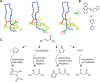
References
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., et al. (2013). Ribosomally Synthesized and post-translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 30 (1), 108–160. 10.1039/C2NP20085F - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

