A Risk Score Model Incorporating Three m6A RNA Methylation Regulators and a Related Network of miRNAs-m6A Regulators-m6A Target Genes to Predict the Prognosis of Patients With Ovarian Cancer
- PMID: 34631700
- PMCID: PMC8495156
- DOI: 10.3389/fcell.2021.703969
A Risk Score Model Incorporating Three m6A RNA Methylation Regulators and a Related Network of miRNAs-m6A Regulators-m6A Target Genes to Predict the Prognosis of Patients With Ovarian Cancer
Abstract
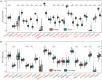
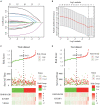
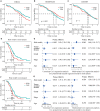
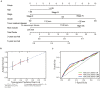
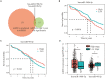
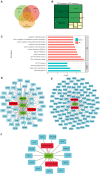
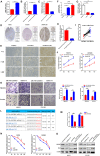
Ovarian cancer (OC) is the leading cause of cancer-related death among all gynecological tumors. N6-methyladenosine (m6A)-related regulators play essential roles in various tumors, including OC. However, the expression of m6A RNA methylation regulators and the related regulatory network in OC and their correlations with prognosis remain largely unknown. In the current study, we obtained the genome datasets of OC from GDC and GTEx database and analyzed the mRNA levels of 21 key m6A regulators in OC and normal human ovarian tissues. The expression levels of 7 m6A regulators were lower in both the OC tissues and the high-stage group. Notably, the 5-year survival rate of patients with OC presenting low VIRMA expression or high HNRNPA2B1 expression was higher than that of the controls. Next, a risk score model based on the three selected m6A regulators (VIRMA, IGF2BP1, and HNRNPA2B1) was built by performing a LASSO regression analysis, and the moderate accuracy of the risk score model to predict the prognosis of patients with OC was examined by performing ROC curve, nomogram, and univariate and multivariate Cox regression analyses. In addition, a regulatory network of miRNAs-m6A regulators-m6A target genes, including 2 miRNAs, 3 m6A regulators, and 47 mRNAs, was constructed, and one of the pathways, namely, miR-196b-5p-IGF2BP1-PTEN, was initially validated based on bioinformatic analysis and assay verification. These results demonstrated that the risk score model composed of three m6A RNA methylation regulators and the related network of miRNAs-m6A regulators-m6A target genes is valuable for predicting the prognosis of patients with OC, and these molecules may serve as potential biomarkers or therapeutic targets in the future.
Keywords: RNA methylation; m6A; ovarian cancer; prognosis; risk model.
Copyright © 2021 Li, Ren, Chen, Yang, Zhang, Wang, Zhu, Li, Yang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
m6A RNA methylation regulators were associated with the malignancy and prognosis of ovarian cancer.Bioengineered. 2021 Dec;12(1):3159-3176. doi: 10.1080/21655979.2021.1946305. Bioengineered. 2021. PMID: 34187307 Free PMC article.
-
m6A RNA methylation regulators play an important role in the prognosis of patients with testicular germ cell tumor.Transl Androl Urol. 2021 Feb;10(2):662-679. doi: 10.21037/tau-20-963. Transl Androl Urol. 2021. PMID: 33718069 Free PMC article.
-
N6-Methyladenosine-Related RNA Signature Predicting the Prognosis of Ovarian Cancer.Recent Pat Anticancer Drug Discov. 2021;16(3):407-416. doi: 10.2174/1574892816666210615164645. Recent Pat Anticancer Drug Discov. 2021. PMID: 34137363
-
RNA m6A methylation regulators in ovarian cancer.Cancer Cell Int. 2021 Nov 18;21(1):609. doi: 10.1186/s12935-021-02318-8. Cancer Cell Int. 2021. PMID: 34794452 Free PMC article. Review.
-
Research Advances in the Roles of N6-Methyladenosine Modification in Ovarian Cancer.Cancer Control. 2024 Jan-Dec;31:10732748241256819. doi: 10.1177/10732748241256819. Cancer Control. 2024. PMID: 38755968 Free PMC article. Review.
Cited by
-
The risk of COVID-19 can be predicted by a nomogram based on m6A-related genes.Infect Genet Evol. 2022 Dec;106:105389. doi: 10.1016/j.meegid.2022.105389. Epub 2022 Nov 29. Infect Genet Evol. 2022. PMID: 36460278 Free PMC article.
-
Emerging role of m6A modification in ovarian cancer: progression, drug resistance, and therapeutic prospects.Front Oncol. 2024 Mar 13;14:1366223. doi: 10.3389/fonc.2024.1366223. eCollection 2024. Front Oncol. 2024. PMID: 38544837 Free PMC article. Review.
-
The development of a novel signature based on the m6A RNA methylation regulator-related ceRNA network to predict prognosis and therapy response in sarcomas.Front Genet. 2022 Oct 12;13:894080. doi: 10.3389/fgene.2022.894080. eCollection 2022. Front Genet. 2022. PMID: 36313417 Free PMC article.
-
Epigenetic regulation in female reproduction: the impact of m6A on maternal-fetal health.Cell Death Discov. 2025 Feb 4;11(1):43. doi: 10.1038/s41420-025-02324-z. Cell Death Discov. 2025. PMID: 39904996 Free PMC article. Review.
-
Roles of N6-methyladenosine (m6A) modifications in gynecologic cancers: mechanisms and therapeutic targeting.Exp Hematol Oncol. 2022 Nov 12;11(1):98. doi: 10.1186/s40164-022-00357-z. Exp Hematol Oncol. 2022. PMID: 36371254 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

