Unraveling a Nosocomial Outbreak of COVID-19: The Role of Whole-Genome Sequence Analysis
- PMID: 34631912
- PMCID: PMC7989189
- DOI: 10.1093/ofid/ofab120
Unraveling a Nosocomial Outbreak of COVID-19: The Role of Whole-Genome Sequence Analysis
Abstract
Background: The coronavirus disease 2019 (COVID-19) pandemic poses many epidemiological challenges. The investigation of nosocomial transmission is usually performed via thorough investigation of an index case and subsequent contact tracing. Notably, this approach has a subjective component, and there is accumulating evidence that whole-genome sequencing of the virus may provide more objective insight.
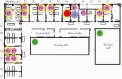
Methods: We report a large nosocomial outbreak in 1 of the medicine departments in our institution. Following intensive epidemiological investigation, we discovered that 1 of the patients involved was suffering from persistent COVID-19 while initially thought to be a recovering patient. She was therefore deemed to be the most likely source of the outbreak. We then performed whole-genome sequencing of the virus of 14 infected individuals involved in the outbreak.
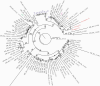
Results: Surprisingly, the results of whole-genome sequencing refuted our initial hypothesis. A phylogenetic tree of the samples showed multiple introductions of the virus into the ward, 1 of which led to a cluster of 10 of the infected individuals. Importantly, the results pointed in the direction of a specific index patient that was different from the 1 that arose from our initial investigation.
Conclusions: These results underscore the important added value of using whole-genome sequencing in epidemiological investigations as it may reveal unexpected connections between cases and aid in understanding transmission dynamics, especially in the setting of a pandemic where multiple possible index cases exist simultaneously.
Keywords: COVID-19; epidemiology; nosocomial; outbreak; sequencing.
© The Author(s) 2021. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Figures

Similar articles
-
Combining epidemiological data and whole genome sequencing to understand SARS-CoV-2 transmission dynamics in a large tertiary care hospital during the first COVID-19 wave in The Netherlands focusing on healthcare workers.Antimicrob Resist Infect Control. 2023 May 10;12(1):46. doi: 10.1186/s13756-023-01247-7. Antimicrob Resist Infect Control. 2023. PMID: 37165456 Free PMC article.
-
Whole genome sequencing reveals a prolonged and spatially spread nosocomial outbreak of Panton-Valentine leucocidin-positive meticillin-resistant Staphylococcus aureus (USA300).J Hosp Infect. 2019 Mar;101(3):327-332. doi: 10.1016/j.jhin.2018.09.007. Epub 2018 Sep 18. J Hosp Infect. 2019. PMID: 30240815
-
Unexpected details regarding nosocomial transmission revealed by whole-genome sequencing of severe acute respiratory coronavirus virus 2 (SARS-CoV-2).Infect Control Hosp Epidemiol. 2022 Oct;43(10):1403-1407. doi: 10.1017/ice.2021.374. Epub 2021 Aug 20. Infect Control Hosp Epidemiol. 2022. PMID: 34496989 Free PMC article.
-
Whole-Genome Sequencing as Tool for Investigating International Tuberculosis Outbreaks: A Systematic Review.Front Public Health. 2019 Apr 17;7:87. doi: 10.3389/fpubh.2019.00087. eCollection 2019. Front Public Health. 2019. PMID: 31058125 Free PMC article.
-
Whole-Genome Sequencing of Bacterial Pathogens: the Future of Nosocomial Outbreak Analysis.Clin Microbiol Rev. 2017 Oct;30(4):1015-1063. doi: 10.1128/CMR.00016-17. Clin Microbiol Rev. 2017. PMID: 28855266 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Outbreak Investigation Using Contact Tracing and Whole-Genome Sequencing in an Ontario Tertiary Care Hospital.Microbiol Spectr. 2023 Jun 15;11(3):e0190022. doi: 10.1128/spectrum.01900-22. Epub 2023 Apr 24. Microbiol Spectr. 2023. PMID: 37093060 Free PMC article.
-
The utility of whole-genome sequencing to inform epidemiologic investigations of SARS-CoV-2 clusters in acute-care hospitals.Infect Control Hosp Epidemiol. 2024 Feb;45(2):144-149. doi: 10.1017/ice.2023.274. Epub 2023 Dec 22. Infect Control Hosp Epidemiol. 2024. PMID: 38130169 Free PMC article.
-
Combining epidemiological data and whole genome sequencing to understand SARS-CoV-2 transmission dynamics in a large tertiary care hospital during the first COVID-19 wave in The Netherlands focusing on healthcare workers.Antimicrob Resist Infect Control. 2023 May 10;12(1):46. doi: 10.1186/s13756-023-01247-7. Antimicrob Resist Infect Control. 2023. PMID: 37165456 Free PMC article.
-
A novel approach to correcting attribution of Clostridioides difficile in a healthcare setting.Antimicrob Steward Healthc Epidemiol. 2023 Dec 20;3(1):e246. doi: 10.1017/ash.2023.516. eCollection 2023. Antimicrob Steward Healthc Epidemiol. 2023. PMID: 38156213 Free PMC article.
-
State-wide genomic epidemiology investigations of COVID-19 in healthcare workers in 2020 Victoria, Australia: Qualitative thematic analysis to provide insights for future pandemic preparedness.Lancet Reg Health West Pac. 2022 Jun 3;25:100487. doi: 10.1016/j.lanwpc.2022.100487. eCollection 2022 Aug. Lancet Reg Health West Pac. 2022. PMID: 35677391 Free PMC article.
References
-
- Oude Munnink BB, Nieuwenhuijse DF, Stein M, et al. ; Dutch-Covid-19 response team . Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med 2020; 26:1405–10. - PubMed

