Cancer cell immune mimicry delineates onco-immunologic modulation
- PMID: 34632332
- PMCID: PMC8487027
- DOI: 10.1016/j.isci.2021.103133
Cancer cell immune mimicry delineates onco-immunologic modulation
Abstract
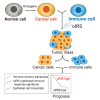
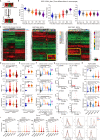
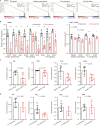
Immune transcripts are essential for depicting onco-immunologic interactions. However, whether cancer cells mimic immune transcripts to reprogram onco-immunologic interaction remains unclear. Here, single-cell transcriptomic analyses of 7,737 normal and 37,476 cancer cells reveal increased immune transcripts in cancer cells. Cells gradually acquire immune transcripts in malignant transformation. Notably, cancer cell-derived immune transcripts contribute to distinct prognoses of immune gene signatures. Optimized immune response signature (oIRS), obtained by excluding cancer-related immune genes from immune gene signatures, and offers a more reliable prognostic value. oIRS reveals that antigen presentation, NK cell killing and T cell signaling are associated with favorable prognosis. Patients with higher oIRS expression are associated with favorable responses to immunotherapy. Indeed, CD83+ cell infiltration, which indicates antigen presentation activity, predicts favorable prognosis in breast cancer. These findings unveil that immune mimicry is a distinct cancer hallmark, providing an example of cancer cell plasticity and a refined view of tumor microenvironment.
Keywords: Biological sciences; Genomics; Immunology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The Landscape of the Tumor Microenvironment in Skin Cutaneous Melanoma Reveals a Prognostic and Immunotherapeutically Relevant Gene Signature.Front Cell Dev Biol. 2021 Oct 1;9:739594. doi: 10.3389/fcell.2021.739594. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34660598 Free PMC article.
-
Immune-related prognostic genes signatures in the tumor microenvironment of sarcoma.Math Biosci Eng. 2021 Mar 5;18(3):2243-2257. doi: 10.3934/mbe.2021113. Math Biosci Eng. 2021. PMID: 33892543
-
Integration of multiple key molecules in lung adenocarcinoma identifies prognostic and immunotherapeutic relevant gene signatures.Int Immunopharmacol. 2020 Jun;83:106477. doi: 10.1016/j.intimp.2020.106477. Epub 2020 Apr 8. Int Immunopharmacol. 2020. PMID: 32278127
-
The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin.Front Immunol. 2018 Aug 15;9:1904. doi: 10.3389/fimmu.2018.01904. eCollection 2018. Front Immunol. 2018. PMID: 30158938 Free PMC article. Review.
-
The Role of Autophagy in Tumor Immunology-Complex Mechanisms That May Be Explored Therapeutically.Front Oncol. 2020 Dec 1;10:603661. doi: 10.3389/fonc.2020.603661. eCollection 2020. Front Oncol. 2020. PMID: 33335860 Free PMC article. Review.
Cited by
-
Exploring glioblastoma stem cell heterogeneity: Immune microenvironment modulation and therapeutic opportunities.Front Oncol. 2022 Sep 21;12:995498. doi: 10.3389/fonc.2022.995498. eCollection 2022. Front Oncol. 2022. PMID: 36212415 Free PMC article. Review.
-
Low tristetraprolin expression activates phenotypic plasticity and primes transition to lethal prostate cancer in mice.J Clin Invest. 2024 Nov 19;135(2):e175680. doi: 10.1172/JCI175680. J Clin Invest. 2024. PMID: 39560993 Free PMC article.
-
Newly identified form of phenotypic plasticity of cancer: immunogenic mimicry.Cancer Metastasis Rev. 2023 Mar;42(1):323-334. doi: 10.1007/s10555-023-10087-1. Epub 2023 Feb 8. Cancer Metastasis Rev. 2023. PMID: 36754910 Free PMC article. Review.
-
Identifying Prognostic Biomarkers Related to m6A Modification and Immune Infiltration in Renal Cell Carcinoma.Genes (Basel). 2022 Nov 7;13(11):2059. doi: 10.3390/genes13112059. Genes (Basel). 2022. PMID: 36360294 Free PMC article.
-
Proteome-based molecular subtyping and therapeutic target prediction in gastric cancer.Mol Oncol. 2024 Jun;18(6):1437-1459. doi: 10.1002/1878-0261.13654. Epub 2024 Apr 16. Mol Oncol. 2024. PMID: 38627210 Free PMC article.
References
-
- Bruni D., Angell H.K., Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer. 2020;20:662–680. - PubMed
LinkOut - more resources
Full Text Sources

