Identification of antibiotic pairs that evade concurrent resistance via a retrospective analysis of antimicrobial susceptibility test results
- PMID: 34632433
- PMCID: PMC8496867
- DOI: 10.1016/s2666-5247(21)00118-x
Identification of antibiotic pairs that evade concurrent resistance via a retrospective analysis of antimicrobial susceptibility test results
Abstract
Background: Some antibiotic pairs display a property known as collateral sensitivity in which the evolution of resistance to one antibiotic increases sensitivity to the other. Alternating between collaterally sensitive antibiotics has been proposed as a sustainable solution to the problem of antibiotic resistance. We aimed to identify antibiotic pairs that could be considered for treatment strategies based on alternating antibiotics.
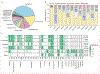
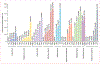
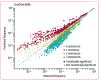
Methods: We did a retrospective analysis of 448 563 antimicrobial susceptibility test results acquired over a 4-year period (Jan 1, 2015, to Dec 31, 2018) from 23 hospitals in the University of Pittsburgh Medical Center (Pittsburgh, PA, USA) hospital system. We used a score based on mutual information to identify pairs of antibiotics displaying disjoint resistance, wherein resistance to one antibiotic is commonly associated with susceptibility to the other and vice versa. We applied this approach to the six most frequently isolated bacterial pathogens (Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Enterococcus faecalis, Pseudomonas aeruginosa, and Proteus mirabilis) and subpopulations of each created by conditioning on resistance to individual antibiotics. To identify higher-order antibiotic interactions, we predicted rates of multidrug resistance for triplets of antibiotics using Markov random fields and compared these to the observed rates.
Findings: We identified 69 antibiotic pairs displaying varying degrees of disjoint resistance for subpopulations of the six bacterial species. However, disjoint resistance was rarely conserved at the species level, with only 6 (0·7%) of 875 antibiotic pairs showing evidence of disjoint resistance. Instead, more than half of antibiotic pairs (465 [53·1%] of 875) exhibited signatures of concurrent resistance, whereby resistance to one antibiotic is associated with resistance to another. We found concurrent resistance to extend to more than two antibiotics, with observed rates of resistance to three antibiotics being higher than predicted from pairwise information alone.
Interpretation: The high frequency of concurrent resistance shows that bacteria have means of counteracting multiple antibiotics at a time. The almost complete absence of disjoint resistance at the species level implies that treatment strategies based on alternating between antibiotics might require subspecies level pathogen identification and be limited to a few antibiotic pairings.
Funding: US National Institutes of Health.
Conflict of interest statement
Declaration of interests We declare no competing interests. Data sharing Individual participant data is not sharable due to University of Pittsburgh institutional review board restrictions. Aggregated resistance data for the pairs and triplets of antibiotics analysed in this study are provided in appendix 2.
Figures


Comment in
-
Identification of antibiotic pairs with disjoint resistance: innovative progress made but questions remain.Lancet Microbe. 2021 Dec;2(12):e651. doi: 10.1016/S2666-5247(21)00274-3. Epub 2021 Oct 28. Lancet Microbe. 2021. PMID: 35544103 No abstract available.
-
Identification of antibiotic pairs with disjoint resistance: innovative progress made but questions remain.Lancet Microbe. 2021 Dec;2(12):e652. doi: 10.1016/S2666-5247(21)00276-7. Epub 2021 Oct 28. Lancet Microbe. 2021. PMID: 35544104 No abstract available.
-
Identification of antibiotic pairs with disjoint resistance: innovative progress made but questions remain - Authors' reply.Lancet Microbe. 2021 Dec;2(12):e653. doi: 10.1016/S2666-5247(21)00281-0. Epub 2021 Oct 28. Lancet Microbe. 2021. PMID: 35544105 No abstract available.
-
No collateral antibiotic sensitivity by alternating antibiotic pairs.Lancet Microbe. 2022 Jan;3(1):e7. doi: 10.1016/S2666-5247(21)00270-6. Epub 2021 Nov 15. Lancet Microbe. 2022. PMID: 35544116 No abstract available.
References
-
- van Duijn PJ, Verbrugghe W, Jorens PG, et al. The effects of antibiotic cycling and mixing on antibiotic resistance in intensive care units: a cluster-randomised crossover trial. Lancet Infect Dis 2018; 18: 401–09. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
