Global approaches for profiling transcription initiation
- PMID: 34632443
- PMCID: PMC8496859
- DOI: 10.1016/j.crmeth.2021.100081
Global approaches for profiling transcription initiation
Abstract
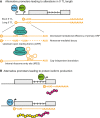
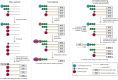
Transcription start site (TSS) selection influences transcript stability and translation as well as protein sequence. Alternative TSS usage is pervasive in organismal development, is a major contributor to transcript isoform diversity in humans, and is frequently observed in human diseases including cancer. In this review, we discuss the breadth of techniques that have been used to globally profile TSSs and the resulting insights into gene regulation, as well as future prospects in this area of inquiry.
Conflict of interest statement
DECLARATION OF INTERESTS R.A.P. and G.E.Z. are employees of eGenesis, Inc. The topics covered in this review are not related to any current work at the company.
Figures
Similar articles
-
Transcription start site-level expression of thyroid transcription factor 1 isoforms in lung adenocarcinoma and its clinicopathological significance.J Pathol Clin Res. 2021 Jul;7(4):361-374. doi: 10.1002/cjp2.213. Epub 2021 May 20. J Pathol Clin Res. 2021. PMID: 34014042 Free PMC article.
-
Alternative transcription start sites contribute to acute-stress-induced transcriptome response in human skeletal muscle.Hum Genomics. 2022 Jul 22;16(1):24. doi: 10.1186/s40246-022-00399-8. Hum Genomics. 2022. PMID: 35869513 Free PMC article.
-
Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis.BMC Genomics. 2011 Oct 14;12:505. doi: 10.1186/1471-2164-12-505. BMC Genomics. 2011. PMID: 21999571 Free PMC article.
-
Regulation of gene expression by alternative promoters.FASEB J. 1996 Mar;10(4):453-60. FASEB J. 1996. PMID: 8647344 Review.
-
Histone variants at the transcription start-site.Trends Genet. 2014 May;30(5):199-209. doi: 10.1016/j.tig.2014.03.002. Epub 2014 Apr 23. Trends Genet. 2014. PMID: 24768041 Review.
Cited by
-
Approaches in Gene Coexpression Analysis in Eukaryotes.Biology (Basel). 2022 Jul 6;11(7):1019. doi: 10.3390/biology11071019. Biology (Basel). 2022. PMID: 36101400 Free PMC article. Review.
-
An improved method for the highly specific detection of transcription start sites.Nucleic Acids Res. 2024 Jan 25;52(2):e7. doi: 10.1093/nar/gkad1116. Nucleic Acids Res. 2024. PMID: 37994784 Free PMC article.
-
From promoter motif to cardiac function: a single DPE motif affects transcription regulation and organ function in vivo.Development. 2024 Jul 15;151(14):dev202355. doi: 10.1242/dev.202355. Epub 2024 Jul 29. Development. 2024. PMID: 38958007 Free PMC article.
-
Transcriptomic Complexity in Strawberry Fruit Development and Maturation Revealed by Nanopore Sequencing.Front Plant Sci. 2022 Jul 13;13:872054. doi: 10.3389/fpls.2022.872054. eCollection 2022. Front Plant Sci. 2022. PMID: 35909727 Free PMC article.
-
Differences in 5'untranslated regions highlight the importance of translational regulation of dosage sensitive genes.Genome Biol. 2024 Apr 29;25(1):111. doi: 10.1186/s13059-024-03248-0. Genome Biol. 2024. PMID: 38685090 Free PMC article.
References
-
- Baldrich P., Tamim S., Mathioni S., Meyers B. Ligation Bias Is a Major Contributor to Nonstoichiometric Abundances of Secondary siRNAs and Impacts Analyses of microRNAs. BioRxiv. 2020 doi: 10.1101/2020.09.14.296616. 2020.09.14.296616. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

