Molecular Diagnoses of X-Linked and Other Genetic Hypophosphatemias: Results From a Sponsored Genetic Testing Program
- PMID: 34633109
- PMCID: PMC9298723
- DOI: 10.1002/jbmr.4454
Molecular Diagnoses of X-Linked and Other Genetic Hypophosphatemias: Results From a Sponsored Genetic Testing Program
Abstract
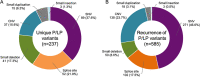
X-linked hypophosphatemia (XLH), a dominant disorder caused by pathogenic variants in the PHEX gene, affects both sexes of all ages and results in elevated serum fibroblast growth factor 23 (FGF23) and below-normal serum phosphate. In XLH, rickets, osteomalacia, short stature, and lower limb deformity may be present with muscle pain and/or weakness/fatigue, bone pain, joint pain/stiffness, hearing difficulty, enthesopathy, osteoarthritis, and dental abscesses. Invitae and Ultragenyx collaborated to provide a no-charge sponsored testing program using a 13-gene next-generation sequencing panel to confirm clinical XLH or aid diagnosis of suspected XLH/other genetic hypophosphatemia. Individuals aged ≥6 months with clinical XLH or suspected genetic hypophosphatemia were eligible. Of 831 unrelated individuals tested between February 2019 and June 2020 in this cross-sectional study, 519 (62.5%) individuals had a pathogenic or likely pathogenic variant in PHEX (PHEX-positive). Among the 312 PHEX-negative individuals, 38 received molecular diagnoses in other genes, including ALPL, CYP27B1, ENPP1, and FGF23; the remaining 274 did not have a molecular diagnosis. Among 319 patients with a provider-reported clinical diagnosis of XLH, 88.7% (n = 283) had a reportable PHEX variant; 81.5% (n = 260) were PHEX-positive. The most common variant among PHEX-positive individuals was an allele with both the gain of exons 13-15 and c.*231A>G (3'UTR variant) (n = 66/519). Importantly, over 80% of copy number variants would have been missed by traditional microarray analysis. A positive molecular diagnosis in 41 probands (4.9%; 29 PHEX positive, 12 non-PHEX positive) resulted in at least one family member receiving family testing. Additional clinical or family member information resulted in variant(s) of uncertain significance (VUS) reclassification to pathogenic/likely pathogenic (P/LP) in 48 individuals, highlighting the importance of segregation and clinical data. In one of the largest XLH genetic studies to date, 65 novel PHEX variants were identified and a high XLH diagnostic yield demonstrated broad insight into the genetic basis of XLH. © 2021 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Keywords: CELL/TISSUE SIGNALING - PARACRINE PATHWAYS - OTHER; DISEASES AND DISORDERS OF/RELATED TO BONE - OTHER; DISORDERS OF CALCIUM/PHOSPHATE METABOLISM- OTHER; GENETIC RESEARCH - HUMAN ASSOCIATION STUDIES; THERAPEUTICS - OTHER.
© 2021 The Authors. Journal of Bone and Mineral Research published by Wiley Periodicals LLC on behalf of American Society for Bone and Mineral Research (ASBMR).
Figures


References
-
- Pavone V, Testa G, Gioitta Iachino S, Evola FR, Avondo S, Sessa G. Hypophosphatemic rickets: etiology, clinical features and treatment. Eur J Orthop Surg Traumatol. 2015;25(2):221‐226. - PubMed
-
- Beck‐Nielsen SS, Brock‐Jacobsen B, Gram J, Brixen K, Jensen TK. Incidence and prevalence of nutritional and hereditary rickets in southern Denmark. Eur J Endocrinol. 2009;160(3):491‐497. - PubMed
-
- Endo I, Fukumoto S, Ozono K, et al. Nationwide survey of fibroblast growth factor 23 (FGF23)‐related hypophosphatemic diseases in Japan: prevalence, biochemical data and treatment. Endocr J. 2015;62(9):811‐816. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

