A pathogenic deletion in Forkhead Box L1 (FOXL1) identifies the first otosclerosis (OTSC) gene
- PMID: 34633540
- PMCID: PMC9034980
- DOI: 10.1007/s00439-021-02381-1
A pathogenic deletion in Forkhead Box L1 (FOXL1) identifies the first otosclerosis (OTSC) gene
Abstract
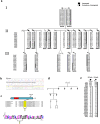
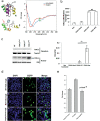
Otosclerosis is a bone disorder of the otic capsule and common form of late-onset hearing impairment. Considered a complex disease, little is known about its pathogenesis. Over the past 20 years, ten autosomal dominant loci (OTSC1-10) have been mapped but no genes identified. Herein, we map a new OTSC locus to a 9.96 Mb region within the FOX gene cluster on 16q24.1 and identify a 15 bp coding deletion in Forkhead Box L1 co-segregating with otosclerosis in a Caucasian family. Pre-operative phenotype ranges from moderate to severe hearing loss to profound sensorineural loss requiring a cochlear implant. Mutant FOXL1 is both transcribed and translated and correctly locates to the cell nucleus. However, the deletion of 5 residues in the C-terminus of mutant FOXL1 causes a complete loss of transcriptional activity due to loss of secondary (alpha helix) structure. FOXL1 (rs764026385) was identified in a second unrelated case on a shared background. We conclude that FOXL1 (rs764026385) is pathogenic and causes autosomal dominant otosclerosis and propose a key inhibitory role for wildtype Foxl1 in bone remodelling in the otic capsule. New insights into the molecular pathology of otosclerosis from this study provide molecular targets for non-invasive therapeutic interventions.
© 2021. The Author(s).
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures

References
-
- Abdelfatah N (2014) The genetic aetiology of otosclerosis in the population of Newfoundland and Labrador. Doctoral dissertation. Memorial University of Newfoundland, Canada
-
- Bel Hadj Ali I, Thys M, Beltaief N, Schrauwen I, Hilgert N, Vanderstraeten K, Dieltjens N, Mnif E, Hachicha S, Besbes G, Ben Arab S, Van Camp G. A new locus for otosclerosis, OTSC8, maps to the pericentromeric region of chromosome 9. Hum Genet. 2008;123:267–272. doi: 10.1007/s00439-008-0470-3. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

