Selective sorting of ancestral introgression in maize and teosinte along an elevational cline
- PMID: 34634032
- PMCID: PMC8530355
- DOI: 10.1371/journal.pgen.1009810
Selective sorting of ancestral introgression in maize and teosinte along an elevational cline
Abstract
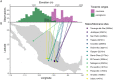
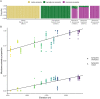
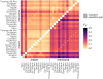
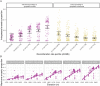
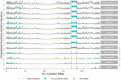
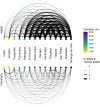
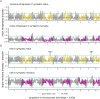
While often deleterious, hybridization can also be a key source of genetic variation and pre-adapted haplotypes, enabling rapid evolution and niche expansion. Here we evaluate these opposing selection forces on introgressed ancestry between maize (Zea mays ssp. mays) and its wild teosinte relative, mexicana (Zea mays ssp. mexicana). Introgression from ecologically diverse teosinte may have facilitated maize's global range expansion, in particular to challenging high elevation regions (> 1500 m). We generated low-coverage genome sequencing data for 348 maize and mexicana individuals to evaluate patterns of introgression in 14 sympatric population pairs, spanning the elevational range of mexicana, a teosinte endemic to the mountains of Mexico. While recent hybrids are commonly observed in sympatric populations and mexicana demonstrates fine-scale local adaptation, we find that the majority of mexicana ancestry tracts introgressed into maize over 1000 generations ago. This mexicana ancestry seems to have maintained much of its diversity and likely came from a common ancestral source, rather than contemporary sympatric populations, resulting in relatively low FST between mexicana ancestry tracts sampled from geographically distant maize populations. Introgressed mexicana ancestry in maize is reduced in lower-recombination rate quintiles of the genome and around domestication genes, consistent with pervasive selection against introgression. However, we also find mexicana ancestry increases across the sampled elevational gradient and that high introgression peaks are most commonly shared among high-elevation maize populations, consistent with introgression from mexicana facilitating adaptation to the highland environment. In the other direction, we find patterns consistent with adaptive and clinal introgression of maize ancestry into sympatric mexicana at many loci across the genome, suggesting that maize also contributes to adaptation in mexicana, especially at the lower end of its elevational range. In sympatric maize, in addition to high introgression regions we find many genomic regions where selection for local adaptation maintains steep gradients in introgressed mexicana ancestry across elevation, including at least two inversions: the well-characterized 14 Mb Inv4m on chromosome 4 and a novel 3 Mb inversion Inv9f surrounding the macrohairless1 locus on chromosome 9. Most outlier loci with high mexicana introgression show no signals of sweeps or local sourcing from sympatric populations and so likely represent ancestral introgression sorted by selection, resulting in correlated but distinct outcomes of introgression in different contemporary maize populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

