Differential pathogenesis of closely related 2018 Nigerian outbreak clade III Lassa virus isolates
- PMID: 34634087
- PMCID: PMC8530337
- DOI: 10.1371/journal.ppat.1009966
Differential pathogenesis of closely related 2018 Nigerian outbreak clade III Lassa virus isolates
Abstract
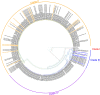
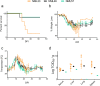
Nigeria continues to experience ever increasing annual outbreaks of Lassa fever (LF). The World Health Organization has recently declared Lassa virus (LASV) as a priority pathogen for accelerated research leading to a renewed international effort to develop relevant animal models of disease and effective countermeasures to reduce LF morbidity and mortality in endemic West African countries. A limiting factor in evaluating medical countermeasures against LF is a lack of well characterized animal models outside of those based on infection with LASV strain Josiah originating form Sierra Leone, circa 1976. Here we genetically characterize five recent LASV isolates collected from the 2018 outbreak in Nigeria. Three isolates were further evaluated in vivo and despite being closely related and from the same spatial / geographic region of Nigeria, only one of the three isolates proved lethal in strain 13 guinea pigs and non-human primates (NHP). Additionally, this isolate exhibited atypical pathogenesis characteristics in the NHP model, most notably respiratory failure, not commonly described in hemorrhagic cases of LF. These results suggest that there is considerable phenotypic heterogeneity in LASV infections in Nigeria, which leads to a multitude of pathogenesis characteristics that could account for differences between subclinical and lethal LF infections. Most importantly, the development of disease models using currently circulating LASV strains in West Africa are critical for the evaluation of potential vaccines and medical countermeasures.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Ogbu O, Ajuluchukwu E, Uneke CJ. Lassa fever in West African sub-region: An overview. Journal of Vector Borne Diseases. 2007. pp. 1–11. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

