The Ensembl COVID-19 resource: ongoing integration of public SARS-CoV-2 data
- PMID: 34634797
- PMCID: PMC8524594
- DOI: 10.1093/nar/gkab889
The Ensembl COVID-19 resource: ongoing integration of public SARS-CoV-2 data
Abstract
The COVID-19 pandemic has seen unprecedented use of SARS-CoV-2 genome sequencing for epidemiological tracking and identification of emerging variants. Understanding the potential impact of these variants on the infectivity of the virus and the efficacy of emerging therapeutics and vaccines has become a cornerstone of the fight against the disease. To support the maximal use of genomic information for SARS-CoV-2 research, we launched the Ensembl COVID-19 browser; the first virus to be encompassed within the Ensembl platform. This resource incorporates a new Ensembl gene set, multiple variant sets, and annotation from several relevant resources aligned to the reference SARS-CoV-2 assembly. Since the first release in May 2020, the content has been regularly updated using our new rapid release workflow, and tools such as the Ensembl Variant Effect Predictor have been integrated. The Ensembl COVID-19 browser is freely available at https://covid-19.ensembl.org.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

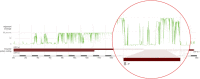
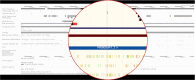
References
-
- Flynn J.A., Purushotham D., Choudhary M.N.K., Zhuo X., Fan C., Matt G., Li D., Wang T.. Exploring the coronavirus pandemic with the WashU Virus Genome Browser. Nat. Genet. 2020; 52:986–991. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

