Efflux Impacts Intracellular Accumulation Only in Actively Growing Bacterial Cells
- PMID: 34634938
- PMCID: PMC8510537
- DOI: 10.1128/mBio.02608-21
Efflux Impacts Intracellular Accumulation Only in Actively Growing Bacterial Cells
Abstract
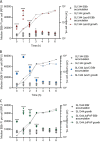
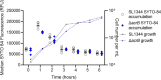
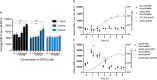
For antibiotics with intracellular targets, effective treatment of bacterial infections requires the drug to accumulate to a high concentration inside cells. Bacteria produce a complex cell envelope and possess drug export efflux pumps to limit drug accumulation inside cells. Decreasing cell envelope permeability and increasing efflux pump activity can reduce intracellular accumulation of antibiotics and are commonly seen in antibiotic-resistant strains. Here, we show that the balance between influx and efflux differs depending on bacterial growth phase in Gram-negative bacteria. Accumulation of the fluorescent compound ethidium bromide (EtBr) was measured in Salmonella enterica serovar Typhimurium SL1344 (wild type) and efflux deficient (ΔacrB) strains during growth. In SL1344, EtBr accumulation remained low, regardless of growth phase, and did not correlate with acrAB transcription. EtBr accumulation in the ΔacrB strains was high in exponential phase but dropped sharply later in growth, with no significant difference from that in SL1344 in stationary phase. Low EtBr accumulation in stationary phase was not due to the upregulation of other efflux pumps but instead was due to decreased permeability of the envelope in stationary phase. Transcriptome sequencing (RNA-seq) identified changes in expression of several pathways that remodel the envelope in stationary phase, leading to lower permeability. IMPORTANCE This study shows that efflux is important for maintaining low intracellular accumulation only in actively growing cells and that envelope permeability is the predominant factor in stationary-phase cells. This conclusion means that (i) antibiotics with intracellular targets may be less effective in complex infections with nongrowing or slow-growing bacteria, where intracellular accumulation may be low; (ii) efflux inhibitors may be successful in potentiating the activity of existing antibiotics, but potentially only for bacterial infections where cells are actively growing; and (iii) the remodeling of the cell envelope prior to stationary phase could provide novel drug targets.
Keywords: antibiotic resistance; efflux pumps; membrane permeability.
Figures



References
-
- Dupont H, Choinier P, Roche D, Adiba S, Sookdeb M, Branger C, Denamur E, Mammeri H. 2017. Structural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25b:H4 sequence type 131 clinical isolate. Antimicrob Agents Chemother 61:e00014-17. doi:10.1128/AAC.00014-17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
