Identification of common microRNA between COPD and non-small cell lung cancer through pathway enrichment analysis
- PMID: 34635059
- PMCID: PMC8507163
- DOI: 10.1186/s12863-021-00986-z
Identification of common microRNA between COPD and non-small cell lung cancer through pathway enrichment analysis
Abstract
Background: Different factors have been introduced which influence the pathogenesis of chronic obstructive pulmonary disease (COPD) and non-small cell lung cancer (NSCLC). COPD as an independent factor is involved in the development of lung cancer. Moreover, there are certain resemblances between NSCLC and COPD, such as growth factors, activation of intracellular pathways, as well as epigenetic factors. One of the best approaches to understand the possible shared pathogenesis routes between COPD and NSCLC is to study the biological pathways that are activated. MicroRNAs (miRNAs) are critical biomolecules that implicate the regulation of several biological and cellular processes. As such, the main goal of this study was to use a systems biology approach to discover common dysregulated miRNAs between COPD and NSCLC, one that targets most genes within common enriched pathways.
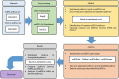
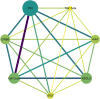
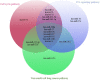
Results: To reconstruct the miRNA-pathways for each disease, we used the microarray miRNA expression data. Then, we employed "miRNA set enrichment analysis" (MiRSEA) to identify the most significant joint miRNAs between COPD and NSCLC based on the enrichment scores. Overall, our study revealed the involvement of the targets of miRNAs (such as has-miR-15b, hsa-miR-106a, has-miR-17, has-miR-103, and has-miR-107) in the most important common biological pathways.
Conclusions: According to the promising results of the pathway analysis, the identified miRNAs can be utilized as the new potential signatures for therapy through understanding the molecular mechanisms of both diseases.
Keywords: COPD; Non-small cell lung Cancer; Pathway analysis; miRNA.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer.Oncotarget. 2016 Jan 26;7(4):4611-23. doi: 10.18632/oncotarget.6566. Oncotarget. 2016. PMID: 26672767 Free PMC article.
-
Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer.BMC Cancer. 2020 Apr 16;20(1):329. doi: 10.1186/s12885-020-06829-x. BMC Cancer. 2020. PMID: 32299382 Free PMC article.
-
Gene expression networks in COPD: microRNA and mRNA regulation.Thorax. 2012 Feb;67(2):122-31. doi: 10.1136/thoraxjnl-2011-200089. Epub 2011 Sep 22. Thorax. 2012. PMID: 21940491
-
MicroRNAs in non-small cell lung cancer and idiopathic pulmonary fibrosis.J Hum Genet. 2017 Jan;62(1):57-65. doi: 10.1038/jhg.2016.98. Epub 2016 Aug 4. J Hum Genet. 2017. PMID: 27488441 Review.
-
Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients.Curr Probl Cancer. 2020 Aug;44(4):100540. doi: 10.1016/j.currproblcancer.2020.100540. Epub 2020 Jan 23. Curr Probl Cancer. 2020. PMID: 32007320 Review.
Cited by
-
MicroRNA Monitoring in Human Alveolar Macrophages from Patients with Smoking-Related Lung Diseases: A Preliminary Study.Biomedicines. 2024 May 9;12(5):1050. doi: 10.3390/biomedicines12051050. Biomedicines. 2024. PMID: 38791013 Free PMC article.
-
Biological and Genetic Mechanisms of COPD, Its Diagnosis, Treatment, and Relationship with Lung Cancer.Biomedicines. 2023 Feb 3;11(2):448. doi: 10.3390/biomedicines11020448. Biomedicines. 2023. PMID: 36830984 Free PMC article. Review.
-
Lung Cancer in the Course of COPD-Emerging Problems Today.Cancers (Basel). 2022 Aug 6;14(15):3819. doi: 10.3390/cancers14153819. Cancers (Basel). 2022. PMID: 35954482 Free PMC article. Review.
-
Impact of chronic obstructive pulmonary disease on the efficacy and safety of neoadjuvant immune checkpoint inhibitors combined with chemotherapy for resectable non-small cell lung cancer: a retrospective cohort study.BMC Cancer. 2024 Jan 30;24(1):153. doi: 10.1186/s12885-024-11902-w. BMC Cancer. 2024. PMID: 38291354 Free PMC article.
-
Altered Differentiation and Inflammation Profiles Contribute to Enhanced Innate Responses in Severe COPD Epithelium to Rhinovirus Infection.Front Med (Lausanne). 2022 Feb 25;9:741989. doi: 10.3389/fmed.2022.741989. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35280870 Free PMC article.
References
-
- Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, Chen R, Decramer M, Fabbri LM, Frith P, Halpin DMG, López Varela MV, Nishimura M, Roche N, Rodriguez-Roisin R, Sin DD, Singh D, Stockley R, Vestbo J, Wedzicha JA, Agustí A. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. Am J Respir Crit Care Med. 2017;195(5):557–582. doi: 10.1164/rccm.201701-0218PP. - DOI - PubMed
-
- Butler SJ, Ellerton L, Goldstein RS, Brooks D. Prevalence of lung cancer in chronic obstructive pulmonary disease: a systematic review. Respir Med X. 2019;1:100003. doi: 10.1016/j.yrmex.2019.100003. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
