A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS)
- PMID: 34635859
- PMCID: PMC10824140
- DOI: 10.1038/s41596-021-00593-3
A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS)
Abstract
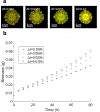
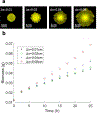
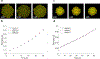
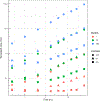
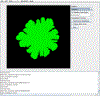
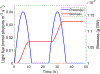
Genome-scale stoichiometric modeling of metabolism has become a standard systems biology tool for modeling cellular physiology and growth. Extensions of this approach are emerging as a valuable avenue for predicting, understanding and designing microbial communities. Computation of microbial ecosystems in time and space (COMETS) extends dynamic flux balance analysis to generate simulations of multiple microbial species in molecularly complex and spatially structured environments. Here we describe how to best use and apply the most recent version of COMETS, which incorporates a more accurate biophysical model of microbial biomass expansion upon growth, evolutionary dynamics and extracellular enzyme activity modules. In addition to a command-line option, COMETS includes user-friendly Python and MATLAB interfaces compatible with the well-established COBRA models and methods, as well as comprehensive documentation and tutorials. This protocol provides a detailed guideline for installing, testing and applying COMETS to different scenarios, generating simulations that take from a few minutes to several days to run, with broad applicability to microbial communities across biomes and scales.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare that they have no competing financial interests.
Figures


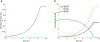

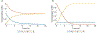



References
-
- Vorholt JA, Vogel C, Carlström CI & Müller DB Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 22, 142–155 (2017). - PubMed

