Palmdelphin Regulates Nuclear Resilience to Mechanical Stress in the Endothelium
- PMID: 34636652
- PMCID: PMC8589083
- DOI: 10.1161/CIRCULATIONAHA.121.054182
Palmdelphin Regulates Nuclear Resilience to Mechanical Stress in the Endothelium
Abstract
Background: PALMD (palmdelphin) belongs to the family of paralemmin proteins implicated in cytoskeletal regulation. Single nucleotide polymorphisms in the PALMD locus that result in reduced expression are strong risk factors for development of calcific aortic valve stenosis and predict severity of the disease.
Methods: Immunodetection and public database screening showed dominant expression of PALMD in endothelial cells (ECs) in brain and cardiovascular tissues including aortic valves. Mass spectrometry, coimmunoprecipitation, and immunofluorescent staining allowed identification of PALMD partners. The consequence of loss of PALMD expression was assessed in small interferring RNA-treated EC cultures, knockout mice, and human valve samples. RNA sequencing of ECs and transcript arrays on valve samples from an aortic valve study cohort including patients with the single nucleotide polymorphism rs7543130 informed about gene regulatory changes.
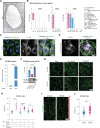
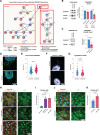
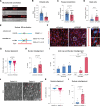
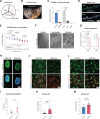
Results: ECs express the cytosolic PALMD-KKVI splice variant, which associated with RANGAP1 (RAN GTP hydrolyase activating protein 1). RANGAP1 regulates the activity of the GTPase RAN and thereby nucleocytoplasmic shuttling via XPO1 (Exportin1). Reduced PALMD expression resulted in subcellular relocalization of RANGAP1 and XPO1, and nuclear arrest of the XPO1 cargoes p53 and p21. This indicates an important role for PALMD in nucleocytoplasmic transport and consequently in gene regulation because of the effect on localization of transcriptional regulators. Changes in EC responsiveness on loss of PALMD expression included failure to form a perinuclear actin cap when exposed to flow, indicating lack of protection against mechanical stress. Loss of the actin cap correlated with misalignment of the nuclear long axis relative to the cell body, observed in PALMD-deficient ECs, Palmd-/- mouse aorta, and human aortic valve samples derived from patients with calcific aortic valve stenosis. In agreement with these changes in EC behavior, gene ontology analysis showed enrichment of nuclear- and cytoskeleton-related terms in PALMD-silenced ECs.
Conclusions: We identify RANGAP1 as a PALMD partner in ECs. Disrupting the PALMD/RANGAP1 complex alters the subcellular localization of RANGAP1 and XPO1, and leads to nuclear arrest of the XPO1 cargoes p53 and p21, accompanied by gene regulatory changes and loss of actin-dependent nuclear resilience. Combined, these consequences of reduced PALMD expression provide a mechanistic underpinning for PALMD's contribution to calcific aortic valve stenosis pathology.
Keywords: aortic valve stenosis; endothelial cells; nucleocytoplasmic transport; palmdelphin.
Figures


References
-
- Hu B, Petrasch-Parwez E, Laue MM, Kilimann MW. Molecular characterization and immunohistochemical localization of palmdelphin, a cytosolic isoform of the paralemmin protein family implicated in membrane dynamics. Eur J Cell Biol. 2005; 84:853–866. doi: 10.1016/j.ejcb.2005.07.002 - PubMed
-
- Kalebic N, Gilardi C, Stepien B, Wilsch-Bräuninger M, Long KR, Namba T, Florio M, Langen B, Lombardot B, Shevchenko A, et al. Neocortical expansion due to increased proliferation of basal progenitors is linked to changes in their morphology. Cell Stem Cell. 2019; 24:535–550.e9. doi: 10.1016/j.stem.2019.02.017 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

