A new prognostic risk model based on autophagy-related genes in kidney renal clear cell carcinoma
- PMID: 34636718
- PMCID: PMC8806698
- DOI: 10.1080/21655979.2021.1976050
A new prognostic risk model based on autophagy-related genes in kidney renal clear cell carcinoma
Abstract
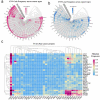
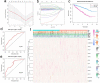
This study aimed to explore the potential role of autophagy-related genes in kidney renal clear cell carcinoma (KIRC) and develop a new prognostic-related risk model. In our research, we used multiple bioinformatics methods to perform a pan-cancer analysis of the CNV, SNV, mRNA expression, and overall survival of autophagy-related genes, and displayed the results in the form of heat maps. We then performed cluster analysis and LASSO regression analysis on these autophagy-related genes in KIRC. In the cluster analysis, we successfully divided patients with KIRC into five clusters and found that there was a clear correlation between the classification and two clinicopathological features: tumor, and stage. In LASSO regression analysis, we used 13 genes to create a new prognostic-related risk model in KIRC. The model showed that the survival rate of patients with KIRC in the high-risk group was significantly lower than that in the low-risk group, and that there was a correlation between this grouping and the patients' metastasis, tumor, stage, grade, and fustat. The results of the ROC curve suggested that this model has good prediction accuracy. The results of multivariate Cox analysis show that the risk score of this model can be used as an independent risk factor for patients with KIRC. In summary, we believe that this research provides valuable data supporting future clinical treatment and scientific research.
Keywords: Autophagy; kidney renal clear cell carcinoma; nomogram; prognostic risk model; tcga.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





References
-
- Levine B, Klionsky DJ.. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. - PubMed
-
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. - PubMed
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–132. - PubMed
-
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
