MCRL: using a reference library to compress a metagenome into a non-redundant list of sequences, considering viruses as a case study
- PMID: 34636854
- PMCID: PMC10060711
- DOI: 10.1093/bioinformatics/btab703
MCRL: using a reference library to compress a metagenome into a non-redundant list of sequences, considering viruses as a case study
Abstract
Motivation: Metagenomes offer a glimpse into the total genomic diversity contained within a sample. Currently, however, there is no straightforward way to obtain a non-redundant list of all putative homologs of a set of reference sequences present in a metagenome.
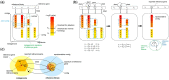
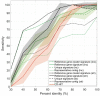
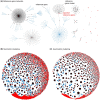
Results: To address this problem, we developed a novel clustering approach called 'metagenomic clustering by reference library' (MCRL), where a reference library containing a set of reference genes is clustered with respect to an assembled metagenome. According to our proposed approach, reference genes homologous to similar sets of metagenomic sequences, termed 'signatures', are iteratively clustered in a greedy fashion, retaining at each step the reference genes yielding the lowest E values, and terminating when signatures of remaining reference genes have a minimal overlap. The outcome of this computation is a non-redundant list of reference genes homologous to minimally overlapping sets of contigs, representing potential candidates for gene families present in the metagenome. Unlike metagenomic clustering methods, there is no need for contigs to overlap to be associated with a cluster, enabling MCRL to draw on more information encoded in the metagenome when computing tentative gene families. We demonstrate how MCRL can be used to extract candidate viral gene families from an oral metagenome and an oral virome that otherwise could not be determined using standard approaches. We evaluate the sensitivity, accuracy and robustness of our proposed method for the viral case study and compare it with existing analysis approaches.
Availability and implementation: https://github.com/a-tadmor/MCRL.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Arnold K. et al. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195–201. - PubMed

