Transcriptomic in silico analysis of bovine Escherichia coli mastitis highlights its immune-related expressed genes as an effective biomarker
- PMID: 34637035
- PMCID: PMC8511192
- DOI: 10.1186/s43141-021-00235-x
Transcriptomic in silico analysis of bovine Escherichia coli mastitis highlights its immune-related expressed genes as an effective biomarker
Abstract
Background: Mastitis is one of the major diseases causing economic loss to the dairy industry by reducing the quantity and quality of milk. Thus, the objective of this scientific study was to find new biomarkers based on genes for the early prediction before its severity.
Methods: In the present study, advanced bioinformatics including hierarchical clustering, enrichment analysis, active site prediction, epigenetic analysis, functional domain identification, and protein docking were used to analyze the important genes that could be utilized as biomarkers and therapeutic targets for mastitis.
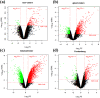
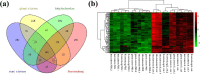
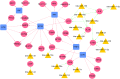
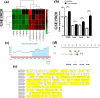
Results: Four differentially expressed genes (DEGs) were identified in different regions of the mammary gland (teat cistern, gland cistern, lobuloalveolar, and Furstenberg's rosette) that resulted in 453, 597, 577, and 636 DEG, respectively. Also, 101 overlapped genes were found by comparing 27 different expressed genes. These genes were associated with eight immune response pathways including NOD-like receptor signaling pathway (IL8, IL18, IL1B, PYDC1) and chemokine signaling pathway (PTK2, IL8, NCF1, CCR1, HCK). Meanwhile, 241 protein-protein interaction networks were developed among overlapped genes. Fifty-seven regulatory events were found between miRNAs, expressed genes, and the transcription factors (TFs) through micro-RNA and transcription factors (miRNA-DEG-TF) regulatory network. The 3D structure docking model of the expressed genes proteins identified their active sites and the binding ligands that could help in choosing the appropriate feed or treatment for affected animals.
Conclusions: The novelty of the distinguished DEG and their pathways in this study is that they can precisely improve the detection biomarkers and treatments techniques of cows' Escherichia coli mastitis disease due to their high affinity with the target site of the mammary gland before appearing the symptoms.
Keywords: Dairy cow; Differentially expressed genes; Escherichia coli; Mastitis; Microarray; Transcriptomic analysis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that the work described was an original manuscript that followed all the ethical regulations and that has not been published previously. Also, no animal or human trails were constructed and no conflict of interest exists in the submission of this manuscript, and the manuscript is approved by all authors to be published.
The authors declare that they have no competing interests.
Figures

Similar articles
-
Transcriptome profiling of Staphylococci-infected cow mammary gland parenchyma.BMC Vet Res. 2017 Jun 6;13(1):161. doi: 10.1186/s12917-017-1088-2. BMC Vet Res. 2017. PMID: 28587645 Free PMC article.
-
Identification of Key Candidate Genes in Dairy Cow in Response to Escherichia coli Mastitis by Bioinformatical Analysis.Front Genet. 2019 Dec 6;10:1251. doi: 10.3389/fgene.2019.01251. eCollection 2019. Front Genet. 2019. PMID: 31921295 Free PMC article.
-
Comparison of microRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus aureus and Escherichia coli.Int J Biol Sci. 2018 Jan 11;14(1):87-99. doi: 10.7150/ijbs.22498. eCollection 2018. Int J Biol Sci. 2018. PMID: 29483828 Free PMC article.
-
Severity of E. coli mastitis is mainly determined by cow factors.Vet Res. 2003 Sep-Oct;34(5):521-64. doi: 10.1051/vetres:2003023. Vet Res. 2003. PMID: 14556694 Review.
-
Maintaining Optimal Mammary Gland Health and Prevention of Mastitis.Front Vet Sci. 2021 Feb 17;8:607311. doi: 10.3389/fvets.2021.607311. eCollection 2021. Front Vet Sci. 2021. PMID: 33681324 Free PMC article. Review.
Cited by
-
Elucidating ascorbate and aldarate metabolism pathway characteristics via integration of untargeted metabolomics and transcriptomics of the kidney of high-fat diet-fed obese mice.PLoS One. 2024 Apr 11;19(4):e0300705. doi: 10.1371/journal.pone.0300705. eCollection 2024. PLoS One. 2024. PMID: 38603672 Free PMC article.
-
Molecular Genomic Study of Inhibin Molecule Production through Granulosa Cell Gene Expression in Inhibin-Deficient Mice.Molecules. 2022 Aug 30;27(17):5595. doi: 10.3390/molecules27175595. Molecules. 2022. PMID: 36080362 Free PMC article.
-
Animal Wellness: The Power of Multiomics and Integrative Strategies: Multiomics in Improving Animal Health.Vet Med Int. 2024 Oct 24;2024:4125118. doi: 10.1155/2024/4125118. eCollection 2024. Vet Med Int. 2024. PMID: 39484643 Free PMC article. Review.
References
-
- Pathak S, Grillo AR, Scarpa M, Brun P, D’Incà R, Nai L, Banerjee A, Cavallo D, Barzon L, Palù G, Sturniolo GC, Buda A, Castagliuolo I. MiR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol Med. 2015;47(5):e164. doi: 10.1038/emm.2015.21. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
