Role of "dual-personality" fragments in HEV adaptation-analysis of Y-domain region
- PMID: 34637041
- PMCID: PMC8511232
- DOI: 10.1186/s43141-021-00238-8
Role of "dual-personality" fragments in HEV adaptation-analysis of Y-domain region
Abstract
Background: Hepatitis E is a liver disease caused by the pathogen hepatitis E virus (HEV). The largest polyprotein open reading frame 1 (ORF1) contains a nonstructural Y-domain region (YDR) whose activity in HEV adaptation remains uncharted. The specific role of disordered regions in several nonstructural proteins has been demonstrated to participate in the multiplication and multiple regulatory functions of the viruses. Thus, intrinsic disorder of YDR including its structural and functional annotation was comprehensively studied by exploiting computational methodologies to delineate its role in viral adaptation.
Results: Based on our findings, it was evident that YDR contains significantly higher levels of ordered regions with less prevalence of disordered residues. Sequence-based analysis of YDR revealed it as a "dual personality" (DP) protein due to the presence of both structured and unstructured (intrinsically disordered) regions. The evolution of YDR was shaped by pressures that lead towards predominance of both disordered and regularly folded amino acids (Ala, Arg, Gly, Ile, Leu, Phe, Pro, Ser, Tyr, Val). Additionally, the predominance of characteristic DP residues (Thr, Arg, Gly, and Pro) further showed the order as well as disorder characteristic possessed by YDR. The intrinsic disorder propensity analysis of YDR revealed it as a moderately disordered protein. All the YDR sequences consisted of molecular recognition features (MoRFs), i.e., intrinsic disorder-based protein-protein interaction (PPI) sites, in addition to several nucleotide-binding sites. Thus, the presence of molecular recognition (PPI, RNA binding, and DNA binding) signifies the YDR's interaction with specific partners, host membranes leading to further viral infection. The presence of various disordered-based phosphorylation sites further signifies the role of YDR in various biological processes. Furthermore, functional annotation of YDR revealed it as a multifunctional-associated protein, due to its susceptibility in binding to a wide range of ligands and involvement in various catalytic activities.
Conclusions: As DP are targets for regulation, thus, YDR contributes to cellular signaling processes through PPIs. As YDR is incompletely understood, therefore, our data on disorder-based function could help in better understanding its associated functions. Collectively, our novel data from this comprehensive investigation is the first attempt to delineate YDR role in the regulation and pathogenesis of HEV.
Keywords: Molecular function; Nucleotide-binding propensity; Phosphorylation; Protein disorder; Protein structure; Protein-binding propensity; Y-domain region (YDR).
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
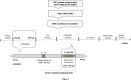
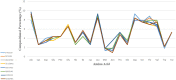
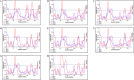
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
