MouseVenue3D: A Markerless Three-Dimension Behavioral Tracking System for Matching Two-Photon Brain Imaging in Free-Moving Mice
- PMID: 34637091
- PMCID: PMC8975979
- DOI: 10.1007/s12264-021-00778-6
MouseVenue3D: A Markerless Three-Dimension Behavioral Tracking System for Matching Two-Photon Brain Imaging in Free-Moving Mice
Abstract
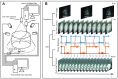
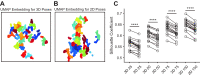
Understanding the connection between brain and behavior in animals requires precise monitoring of their behaviors in three-dimensional (3-D) space. However, there is no available three-dimensional behavior capture system that focuses on rodents. Here, we present MouseVenue3D, an automated and low-cost system for the efficient capture of 3-D skeleton trajectories in markerless rodents. We improved the most time-consuming step in 3-D behavior capturing by developing an automatic calibration module. Then, we validated this process in behavior recognition tasks, and showed that 3-D behavioral data achieved higher accuracy than 2-D data. Subsequently, MouseVenue3D was combined with fast high-resolution miniature two-photon microscopy for synchronous neural recording and behavioral tracking in the freely-moving mouse. Finally, we successfully decoded spontaneous neuronal activity from the 3-D behavior of mice. Our findings reveal that subtle, spontaneous behavior modules are strongly correlated with spontaneous neuronal activity patterns.
Keywords: 3-D pose estimation; Automatic calibration; Behavioral and neural recording; Computational neuroethology; Multi-view cameras.
© 2021. Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences.
Conflict of interest statement
All authors claim that there are no conflicts of interest.
Figures





Similar articles
-
Improved 3D tracking and automated classification of rodents' behavioral activity using depth-sensing cameras.Behav Res Methods. 2020 Oct;52(5):2156-2167. doi: 10.3758/s13428-020-01381-9. Behav Res Methods. 2020. PMID: 32232737
-
Automated markerless pose estimation in freely moving macaques with OpenMonkeyStudio.Nat Commun. 2020 Sep 11;11(1):4560. doi: 10.1038/s41467-020-18441-5. Nat Commun. 2020. PMID: 32917899 Free PMC article.
-
Millimeter field-of-view miniature two-photon microscopy for brain imaging in freely moving mice.Opt Express. 2023 Sep 25;31(20):32925-32934. doi: 10.1364/OE.492674. Opt Express. 2023. PMID: 37859084
-
Mapping brain function in freely moving subjects.Neurosci Biobehav Rev. 2004 Sep;28(5):449-61. doi: 10.1016/j.neubiorev.2004.06.007. Neurosci Biobehav Rev. 2004. PMID: 15465134 Free PMC article. Review.
-
Beyond the three-chamber test: toward a multimodal and objective assessment of social behavior in rodents.Mol Autism. 2022 Oct 25;13(1):41. doi: 10.1186/s13229-022-00521-6. Mol Autism. 2022. PMID: 36284353 Free PMC article. Review.
Cited by
-
Subtle behavioral alterations in the spontaneous behaviors of MPTP mouse model of Parkinson's disease.Transl Psychiatry. 2025 Apr 3;15(1):119. doi: 10.1038/s41398-025-03312-8. Transl Psychiatry. 2025. PMID: 40175343 Free PMC article.
-
Multimodal Nature of the Single-cell Primate Brain Atlas: Morphology, Transcriptome, Electrophysiology, and Connectivity.Neurosci Bull. 2024 Apr;40(4):517-532. doi: 10.1007/s12264-023-01160-4. Epub 2024 Jan 9. Neurosci Bull. 2024. PMID: 38194157 Free PMC article. Review.
-
Mapping the Behavioral Signatures of Shank3b Mice in Both Sexes.Neurosci Bull. 2024 Sep;40(9):1299-1314. doi: 10.1007/s12264-024-01237-8. Epub 2024 Jun 20. Neurosci Bull. 2024. PMID: 38900384 Free PMC article.
-
Anti-drift pose tracker (ADPT), a transformer-based network for robust animal pose estimation cross-species.Elife. 2025 May 6;13:RP95709. doi: 10.7554/eLife.95709. Elife. 2025. PMID: 40326557 Free PMC article.
-
InteBOMB: Integrating generic object tracking and segmentation with pose estimation for animal behavior analysis.Zool Res. 2025 Mar 18;46(2):355-369. doi: 10.24272/j.issn.2095-8137.2024.268. Zool Res. 2025. PMID: 40049663 Free PMC article.
References
-
- Mathis MW, Mathis A. Deep learning tools for the measurement of animal behavior in neuroscience. Curr Opin Neurobiol. 2020;60:1–11. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

