Identification and characterization of SARS-CoV-2 clusters in the EU/EEA in the first pandemic wave: additional elements to trace the route of the virus
- PMID: 34637920
- PMCID: PMC8501518
- DOI: 10.1016/j.meegid.2021.105108
Identification and characterization of SARS-CoV-2 clusters in the EU/EEA in the first pandemic wave: additional elements to trace the route of the virus
Abstract
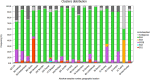
A high-quality dataset of 3289 complete SARS-CoV-2 genomes collected in Europe and European Economic Area (EAA) in the early phase of the first wave of the pandemic was analyzed. Among all single nucleotide mutations, 41 had a frequency ≥ 1%, and the phylogenetic analysis showed at least 6 clusters with a specific mutational profile. These clusters were differentially distributed in the EU/EEA, showing a statistically significant association with the geographic origin. The analysis highlighted that the mutations C14408T and C14805T played an important role in clusters selection and further virus spread. Moreover, the molecular analysis suggests that the SARS-CoV-2 strain responsible for the first Italian confirmed COVID-19 case was already circulating outside the country.
Keywords: COVID-19; Cluster analysis; SARS-CoV-2; SNVs analysis.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
None.
Figures



References
-
- Addetia A., Xie H., Roychoudhury P., Shrestha L., Loprieno M., Huang M.L., Jerome K.R., Greninger A.L. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 2020 Aug;129:104523. doi: 10.1016/j.jcv.2020.104523. - DOI - PMC - PubMed
-
- Albarello F., Pianura E., Di Stefano F., Cristofaro M., Petrone A., Marchioni L., Palazzolo C., Schininà V., Nicastri E., Petrosillo N., Campioni P., Eskild P., Zumla A. Ippolito G; COVID 19 INMI study group. 2019-novel coronavirus severe adult respiratory distress syndrome in two cases in Italy: an uncommon radiological presentation. Int. J. Infect. Dis. 2020 Apr;93:192–197. doi: 10.1016/j.ijid.2020.02.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

