The CADM1 tumor suppressor gene is a major candidate gene in MDS with deletion of the long arm of chromosome 11
- PMID: 34638130
- PMCID: PMC8791575
- DOI: 10.1182/bloodadvances.2021005311
The CADM1 tumor suppressor gene is a major candidate gene in MDS with deletion of the long arm of chromosome 11
Abstract
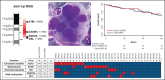
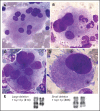
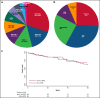
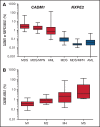
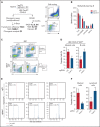
Myelodysplastic syndromes (MDS) represent a heterogeneous group of clonal hematopoietic stem cell disorders characterized by ineffective hematopoiesis leading to peripheral cytopenias and in a substantial proportion of cases to acute myeloid leukemia. The deletion of the long arm of chromosome 11, del(11q), is a rare but recurrent clonal event in MDS. Here, we detail the largest series of 113 cases of MDS and myelodysplastic syndromes/myeloproliferative neoplasms (MDS/MPN) harboring a del(11q) analyzed at clinical, cytological, cytogenetic, and molecular levels. Female predominance, a survival prognosis similar to other MDS, a low monocyte count, and dysmegakaryopoiesis were the specific clinical and cytological features of del(11q) MDS. In most cases, del(11q) was isolated, primary and interstitial encompassing the 11q22-23 region containing ATM, KMT2A, and CBL genes. The common deleted region at 11q23.2 is centered on an intergenic region between CADM1 (also known as Tumor Suppressor in Lung Cancer 1) and NXPE2. CADM1 was expressed in all myeloid cells analyzed in contrast to NXPE2. At the functional level, the deletion of Cadm1 in murine Lineage-Sca1+Kit+ cells modifies the lymphoid-to-myeloid ratio in bone marrow, although not altering their multilineage hematopoietic reconstitution potential after syngenic transplantation. Together with the frequent simultaneous deletions of KMT2A, ATM, and CBL and mutations of ASXL1, SF3B1, and CBL, we show that CADM1 may be important in the physiopathology of the del(11q) MDS, extending its role as tumor-suppressor gene from solid tumors to hematopoietic malignancies.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer; 2008.
-
- WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer; 2017.
-
- Adès L, Itzykson R, Fenaux P. Myelodysplastic syndromes. Lancet. 2014;383(9936):2239-2252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

