Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia-From Molecular Mechanisms to Clinical Relevance
- PMID: 34638304
- PMCID: PMC8508378
- DOI: 10.3390/cancers13194820
Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia-From Molecular Mechanisms to Clinical Relevance
Abstract
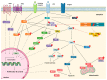
Resistance to targeted therapies is a complex and multifactorial process that culminates in the selection of a cancer clone with the ability to evade treatment. Chronic myeloid leukemia (CML) was the first malignancy recognized to be associated with a genetic alteration, the t(9;22)(q34;q11). This translocation originates the BCR-ABL1 fusion gene, encoding the cytoplasmic chimeric BCR-ABL1 protein that displays an abnormally high tyrosine kinase activity. Although the vast majority of patients with CML respond to Imatinib, a tyrosine kinase inhibitor (TKI), resistance might occur either de novo or during treatment. In CML, the TKI resistance mechanisms are usually subdivided into BCR-ABL1-dependent and independent mechanisms. Furthermore, patients' compliance/adherence to therapy is critical to CML management. Techniques with enhanced sensitivity like NGS and dPCR, the use of artificial intelligence (AI) techniques, and the development of mathematical modeling and computational prediction methods could reveal the underlying mechanisms of drug resistance and facilitate the design of more effective treatment strategies for improving drug efficacy in CML patients. Here we review the molecular mechanisms and other factors involved in resistance to TKIs in CML and the new methodologies to access these mechanisms, and the therapeutic approaches to circumvent TKI resistance.
Keywords: CML; TKI resistance; bioinformatics and artificial intelligence; epigenetics; immune system; new targeted therapies; patient adherence.
Conflict of interest statement
R.A., A.C.G., S.R., I.P.T., J.D.L.R. and A.B.S.R. declare no conflict of interest. A.M.A. has presented at speakers’ bureaus for Novartis, BMS, and Alexion.
Figures

References
-
- Surveillance, Epidemiology, and End Results (SEER) Program SEER*Stat Database: Incidence-SEER Research Data, 9 Registries, Nov 2020 Sub (1975-2018)-Linked To County Attributes-Time Dependent (1990–2018) Income/Rurality, 1969–2019 Counties. National Cancer Institute; Rockville, MD, USA: [(accessed on 25 September 2021)]. DCCPS, Surveillance Research Program. Available online: www.seer.cancer.gov.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

