Single-Tear Proteomics: A Feasible Approach to Precision Medicine
- PMID: 34639092
- PMCID: PMC8509675
- DOI: 10.3390/ijms221910750
Single-Tear Proteomics: A Feasible Approach to Precision Medicine
Abstract
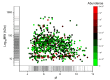
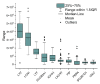
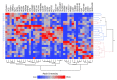
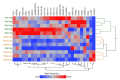
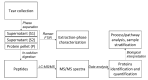
Lacrimal fluid is an attractive source of noninvasive biomarkers, the main limitation being the small sample amounts typically collected. Advanced analytical methods to allow for proteomics profiling from a few microliters are needed to develop innovative biomarkers, with attractive perspectives of applications to precision medicine. This work describes an effective, analytical pipeline for single-tear analysis by ultrahigh-resolution, shotgun proteomics from 23 healthy human volunteers, leading to high-confidence identification of a total of 890 proteins. Highly reproducible quantification was achieved by either peak intensity, peak area, or spectral counting. Hierarchical clustering revealed a stratification of females vs. males that did not emerge from previous studies on pooled samples. Two subjects were monitored weekly over 3 weeks. The samples clustered by withdrawal time of day (morning vs. afternoon) but not by follow-up week, with elevated levels of components of the immune system in the morning samples. This study demonstrates feasibility of single-tear quantitative proteomics, envisaging contributions of this unconventional body fluid to individualized approaches in biomedicine.
Keywords: lacrimal film; liquid biopsies; mass-spectrometry-based proteomics; peripheral body fluids; personalized medicine; single-tear analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lawrenson J.G. 2-anterior eye. In: Efron N., editor. Contact Lens Practice. 3rd ed. Elsevier; Amsterdam, The Netherlands: 2018. pp. 10–27.
-
- Phillips A., Speedwell L. Contact Lenses. 6th ed. Elsevier; Amsterdam, The Netherlands: 2018.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

