NMF-Based Approach for Missing Values Imputation of Mass Spectrometry Metabolomics Data
- PMID: 34641330
- PMCID: PMC8510447
- DOI: 10.3390/molecules26195787
NMF-Based Approach for Missing Values Imputation of Mass Spectrometry Metabolomics Data
Abstract
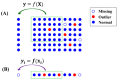
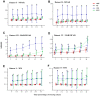
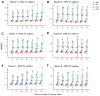
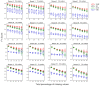
In mass spectrometry (MS)-based metabolomics, missing values (NAs) may be due to different causes, including sample heterogeneity, ion suppression, spectral overlap, inappropriate data processing, and instrumental errors. Although a number of methodologies have been applied to handle NAs, NA imputation remains a challenging problem. Here, we propose a non-negative matrix factorization (NMF)-based method for NA imputation in MS-based metabolomics data, which makes use of both global and local information of the data. The proposed method was compared with three commonly used methods: k-nearest neighbors (kNN), random forest (RF), and outlier-robust (ORI) missing values imputation. These methods were evaluated from the perspectives of accuracy of imputation, retrieval of data structures, and rank of imputation superiority. The experimental results showed that the NMF-based method is well-adapted to various cases of data missingness and the presence of outliers in MS-based metabolic profiles. It outperformed kNN and ORI and showed results comparable with the RF method. Furthermore, the NMF method is more robust and less susceptible to outliers as compared with the RF method. The proposed NMF-based scheme may serve as an alternative NA imputation method which may facilitate biological interpretations of metabolomics data.
Keywords: mass spectrometry; metabolomics data; missing pattern; missing values imputation; non-negative matrix factorization; outliers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
NS-kNN: a modified k-nearest neighbors approach for imputing metabolomics data.Metabolomics. 2018 Nov 23;14(12):153. doi: 10.1007/s11306-018-1451-8. Metabolomics. 2018. PMID: 30830437 Free PMC article.
-
Missing Value Imputation Approach for Mass Spectrometry-based Metabolomics Data.Sci Rep. 2018 Jan 12;8(1):663. doi: 10.1038/s41598-017-19120-0. Sci Rep. 2018. PMID: 29330539 Free PMC article.
-
rMisbeta: A robust missing value imputation approach in transcriptomics and metabolomics data.Comput Biol Med. 2021 Nov;138:104911. doi: 10.1016/j.compbiomed.2021.104911. Epub 2021 Sep 29. Comput Biol Med. 2021. PMID: 34634637
-
Revisiting the Thorny Issue of Missing Values in Single-Cell Proteomics.J Proteome Res. 2023 Sep 1;22(9):2775-2784. doi: 10.1021/acs.jproteome.3c00227. Epub 2023 Aug 2. J Proteome Res. 2023. PMID: 37530557 Review.
-
Machine Learning Applications for Mass Spectrometry-Based Metabolomics.Metabolites. 2020 Jun 13;10(6):243. doi: 10.3390/metabo10060243. Metabolites. 2020. PMID: 32545768 Free PMC article. Review.
Cited by
-
Predicting individual cases of major adolescent psychiatric conditions with artificial intelligence.Transl Psychiatry. 2023 Oct 10;13(1):314. doi: 10.1038/s41398-023-02599-9. Transl Psychiatry. 2023. PMID: 37816706 Free PMC article.
-
RiskPath: Explainable deep learning for multistep biomedical prediction in longitudinal data.medRxiv [Preprint]. 2024 Dec 26:2024.09.19.24313909. doi: 10.1101/2024.09.19.24313909. medRxiv. 2024. PMID: 39371168 Free PMC article. Preprint.
-
Optimizing multi-omics data imputation with NMF and GAN synergy.Bioinformatics. 2024 Nov 1;40(11):btae674. doi: 10.1093/bioinformatics/btae674. Bioinformatics. 2024. PMID: 39546381 Free PMC article.
-
Evaluating Proteomics Imputation Methods with Improved Criteria.J Proteome Res. 2023 Nov 3;22(11):3427-3438. doi: 10.1021/acs.jproteome.3c00205. Epub 2023 Oct 20. J Proteome Res. 2023. PMID: 37861703 Free PMC article.
-
MIRTH: Metabolite Imputation via Rank-Transformation and Harmonization.Genome Biol. 2022 Sep 1;23(1):184. doi: 10.1186/s13059-022-02738-3. Genome Biol. 2022. PMID: 36050754 Free PMC article.
References
-
- Hrydziuszko O., Viant M.R. Missing values in mass spectrometry based metabolomics: An undervalued step in the data processing pipeline. Metabolomics. 2012;8:S161–S174. doi: 10.1007/s11306-011-0366-4. - DOI
-
- Little R.J.A., Rubin D.B. Statistical Analysis with Missing Data. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2002.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

