Probing the Suitability of Different Ca2+ Parameters for Long Simulations of Diisopropyl Fluorophosphatase
- PMID: 34641383
- PMCID: PMC8510429
- DOI: 10.3390/molecules26195839
Probing the Suitability of Different Ca2+ Parameters for Long Simulations of Diisopropyl Fluorophosphatase
Abstract
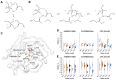
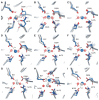
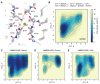
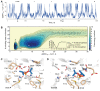
Organophosphate hydrolases are promising as potential biotherapeutic agents to treat poisoning with pesticides or nerve gases. However, these enzymes often need to be further engineered in order to become useful in practice. One example of such enhancement is the alteration of enantioselectivity of diisopropyl fluorophosphatase (DFPase). Molecular modeling techniques offer a unique opportunity to address this task rationally by providing a physical description of the substrate-binding process. However, DFPase is a metalloenzyme, and correct modeling of metal cations is a challenging task generally coming with a tradeoff between simulation speed and accuracy. Here, we probe several molecular mechanical parameter combinations for their ability to empower long simulations needed to achieve a quantitative description of substrate binding. We demonstrate that a combination of the Amber19sb force field with the recently developed 12-6 Ca2+ models allows us to both correctly model DFPase and obtain new insights into the DFP binding process.
Keywords: DFPase; QM/MM; diisopropyl fluorophosphatase; force field; metadynamics; molecular dynamics; substrate binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Matula M., Kucera T., Soukup O., Pejchal J. Enzymatic Degradation of Organophosphorus Pesticides and Nerve Agents by EC: 3.1.8.2. Catalysts. 2020;10:1365. doi: 10.3390/catal10121365. - DOI
-
- Blum M.M., Mustyakimov M., Rüterjans H., Kehe K., Schoenborn B.P., Langan P., Chen J.C.-H. Rapid Determination of Hydrogen Positions and Protonation States of Diisopropyl Fluorophosphatase by Joint Neutron and X-Ray Diffraction Refinement. Proc. Natl. Acad. Sci. USA. 2009;106:713–718. doi: 10.1073/pnas.0807842106. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

