Neurokinin B-Expressing Neurons of the Central Extended Amygdala Mediate Inhibitory Synaptic Input onto Melanin-Concentrating Hormone Neuron Subpopulations
- PMID: 34642212
- PMCID: PMC8612477
- DOI: 10.1523/JNEUROSCI.2600-20.2021
Neurokinin B-Expressing Neurons of the Central Extended Amygdala Mediate Inhibitory Synaptic Input onto Melanin-Concentrating Hormone Neuron Subpopulations
Abstract
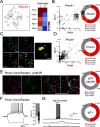
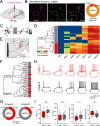
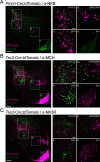
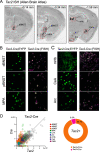
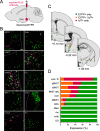
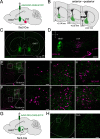
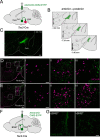
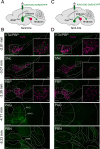
The lateral hypothalamic area (LHA) is a highly conserved brain region critical for maintaining physiological homeostasis and goal-directed behavior. LHA neurons that express melanin-concentrating hormone (MCH) are key regulators of arousal, energy balance, and motivated behavior. However, cellular and functional diversity among LHAMCH neurons is not well understood. Previous anatomic and molecular data suggest that LHAMCH neurons may be parsed into at least two distinct subpopulations, one of which is enriched in neurokinin-3 receptor (NK3R), the receptor for neurokinin B (NKB), encoded by the Tac2 gene. This tachykininergic ligand-receptor system has been implicated in reproduction, fear memory, and stress in other brain regions, but NKB interactions with LHAMCH neurons are poorly understood. We first identified how LHAMCH subpopulations may be distinguished anatomically and electrophysiologically. To dissect functional connectivity between NKB-expressing neurons and LHAMCH neurons, we used Cre-dependent retrograde and anterograde viral tracing in male Tac2-Cre mice and identified Tac2/EYFP+ neurons in the bed nucleus of the stria terminalis and central nucleus of the amygdala, the central extended amygdala, as major sources of NKB input onto LHAMCH neurons. In addition to innervating the LHA, these limbic forebrain NKB neurons also project to midbrain and brainstem targets. Finally, using a dual-virus approach, we found that optogenetic activation of these inputs in slices evokes GABA release onto a subset of LHAMCH neurons but lacked specificity for the NK3R+ subpopulation. Overall, these data define parallel tachykininergic/GABAergic limbic forebrain projections that are positioned to modulate multiple nodes of homeostatic and behavioral control.SIGNIFICANCE STATEMENT The LHA orchestrates fundamental behavioral states in the mammalian hypothalamus, including arousal, energy balance, memory, stress, and motivated behavior. The neuropeptide MCH defines one prominent population of LHA neurons, with multiple roles in the regulation of homeostatic behavior. Outstanding questions remain concerning the upstream inputs that control MCH neurons. We sought to define neurochemically distinct pathways in the mouse brain that may communicate with specific MCH neuron subpopulations using viral-based retrograde and anterograde neural pathway tracing and optogenetics in brain slices. Here, we identify a specific neuropeptide-defined forebrain circuit that makes functional synaptic connections with MCH neuron subpopulations. This work lays the foundation for further manipulating molecularly distinct neural circuits that modulate innate behavioral states.
Keywords: bed nucleus of the stria terminalis; central nucleus of the amygdala; lateral hypothalamic area; melanin-concentrating hormone; neurokinin B; neurokinin-3 receptor.
Copyright © 2021 the authors.
Figures

Similar articles
-
Diencephalic neurons producing melanin-concentrating hormone are influenced by local and multiple extra-hypothalamic tachykininergic projections through the neurokinin 3 receptor.Neuroscience. 2003;119(4):1113-45. doi: 10.1016/s0306-4522(03)00146-5. Neuroscience. 2003. PMID: 12831868
-
Neurochemical Heterogeneity Among Lateral Hypothalamic Hypocretin/Orexin and Melanin-Concentrating Hormone Neurons Identified Through Single-Cell Gene Expression Analysis.eNeuro. 2017 Sep 22;4(5):ENEURO.0013-17.2017. doi: 10.1523/ENEURO.0013-17.2017. eCollection 2017 Sep-Oct. eNeuro. 2017. PMID: 28966976 Free PMC article.
-
Striatal medium spiny neurons terminate in a distinct region in the lateral hypothalamic area and do not directly innervate orexin/hypocretin- or melanin-concentrating hormone-containing neurons.J Neurosci. 2007 Jun 27;27(26):6948-55. doi: 10.1523/JNEUROSCI.0514-07.2007. J Neurosci. 2007. PMID: 17596443 Free PMC article.
-
Melanin-concentrating hormone neuron system: the Wide Web that controls the feeding.Anat Sci Int. 2002 Sep;77(3):149-60. doi: 10.1046/j.0022-7722.2002.00027.x. Anat Sci Int. 2002. PMID: 12422407 Review.
-
Anatomical organization of the melanin-concentrating hormone peptide family in the mammalian brain.Gen Comp Endocrinol. 2011 Jun 1;172(2):185-97. doi: 10.1016/j.ygcen.2011.03.028. Epub 2011 Apr 2. Gen Comp Endocrinol. 2011. PMID: 21463631 Review.
Cited by
-
Melanin concentrating hormone projections to the nucleus accumbens enhance the reward value of food consumption and do not induce feeding or REM sleep.bioRxiv [Preprint]. 2024 Nov 12:2024.11.11.622987. doi: 10.1101/2024.11.11.622987. bioRxiv. 2024. Update in: J Neurosci. 2025 Mar 05;45(10):e1725242024. doi: 10.1523/JNEUROSCI.1725-24.2024. PMID: 39605522 Free PMC article. Updated. Preprint.
-
Central Role of Hypothalamic Circuits for Acupuncture's Anti-Parkinsonian Effects.Adv Sci (Weinh). 2024 Nov;11(43):e2403245. doi: 10.1002/advs.202403245. Epub 2024 Aug 9. Adv Sci (Weinh). 2024. PMID: 39119926 Free PMC article.
-
Electrophysiological properties of melanin-concentrating hormone neuron subpopulations defined by anatomical localization and CART expression.Front Cell Neurosci. 2025 Jan 22;18:1439752. doi: 10.3389/fncel.2024.1439752. eCollection 2024. Front Cell Neurosci. 2025. PMID: 39911950 Free PMC article.
-
Hypothalamic neural circuits regulating energy expenditure.Vitam Horm. 2025;127:79-124. doi: 10.1016/bs.vh.2024.07.004. Epub 2024 Jul 20. Vitam Horm. 2025. PMID: 39864947 Free PMC article. Review.
-
Melanin-Concentrating Hormone Projections to the Nucleus Accumbens Enhance the Reward Value of Food Consumption and Do Not Induce Feeding or REM Sleep.J Neurosci. 2025 Mar 5;45(10):e1725242024. doi: 10.1523/JNEUROSCI.1725-24.2024. J Neurosci. 2025. PMID: 39746823
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
