Schizophrenia-Linked Protein tSNARE1 Regulates Endosomal Trafficking in Cortical Neurons
- PMID: 34642214
- PMCID: PMC8580139
- DOI: 10.1523/JNEUROSCI.0556-21.2021
Schizophrenia-Linked Protein tSNARE1 Regulates Endosomal Trafficking in Cortical Neurons
Abstract
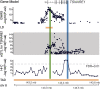
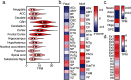
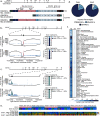
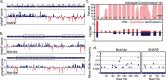
TSNARE1, which encodes the protein tSNARE1, is a high-confidence gene candidate for schizophrenia risk, but nothing is known about its cellular or physiological function. We identified the major gene products of TSNARE1 and their cytoplasmic localization and function in endosomal trafficking in cortical neurons. We validated three primary isoforms of TSNARE1 expressed in human brain, all of which encode a syntaxin-like Qa SNARE domain. RNA-sequencing data from adult and fetal human brain suggested that the majority of tSNARE1 lacks a transmembrane domain that is thought to be necessary for membrane fusion. Biochemical data demonstrate that tSNARE1 can compete with Stx12 for incorporation into an endosomal SNARE complex, supporting its possible role as an inhibitory SNARE. Live-cell imaging in cortical neurons from mice of both sexes demonstrated that brain tSNARE1 isoforms localized to the endosomal network. The most abundant brain isoform, tSNARE1c, localized most frequently to Rab7+ late endosomes, and endogenous tSNARE1 displayed a similar localization in human neural progenitor cells and neuroblastoma cells. In mature rat neurons from both sexes, tSNARE1 localized to the dendritic shaft and dendritic spines, supporting a role for tSNARE1 at the postsynapse. Expression of either tSNARE1b or tSNARE1c, which differ only in their inclusion or exclusion of an Myb-like domain, delayed the trafficking of the dendritic endosomal cargo Nsg1 into late endosomal and lysosomal compartments. These data suggest that tSNARE1 regulates endosomal trafficking in cortical neurons, likely by negatively regulating early endosomal to late endosomal trafficking.SIGNIFICANCE STATEMENT Schizophrenia is a severe and polygenic neuropsychiatric disorder. Understanding the functions of high-confidence candidate genes is critical toward understanding how their dysfunction contributes to schizophrenia pathogenesis. TSNARE1 is one of the high-confidence candidate genes for schizophrenia risk, yet nothing was known about its cellular or physiological function. Here we describe the major isoforms of TSNARE1 and their cytoplasmic localization and function in the endosomal network in cortical neurons. Our results are consistent with the hypothesis that the majority of brain tSNARE1 acts as a negative regulator to endolysosomal trafficking.
Keywords: GWAS; SNARE; endocytosis; late endosome; schizophrenia.
Copyright © 2021 the authors.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
