FBXW11 contributes to stem-cell-like features and liver metastasis through regulating HIC1-mediated SIRT1 transcription in colorectal cancer
- PMID: 34642302
- PMCID: PMC8511012
- DOI: 10.1038/s41419-021-04185-7
FBXW11 contributes to stem-cell-like features and liver metastasis through regulating HIC1-mediated SIRT1 transcription in colorectal cancer
Abstract
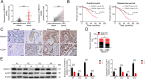
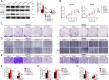
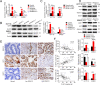
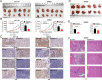
Colorectal tumorigenesis is a heterogeneous disease driven by multiple genetic and epigenetic alterations. F-box and WD repeat domain containing 11 (FBXW11) is a member of the F-box protein family that regulates the ubiquitination of key factors associated with tumor growth and aggressiveness. Our study aimed to explore the role of FBXW11 in the development and metastasis of colorectal cancer (CRC). FBXW11 was overexpressed in colorectal tumor tissues and its overexpression was associated with a poor prognosis of CRC patients. The upregulation of FBXW11 not only promoted cell proliferation, invasion, and migration, but also contributed to maintaining stem-cell features in colorectal tumor cells. Further analysis revealed that FBXW11 targeted hypermethylated in cancer 1 (HIC1) and reduced its stability in CRC cells through ubiquitination. Moreover, the expression of sirtuin 1 (SIRT1), a deacetylase in tumor cells was upregulated by FBXW11 via regulating HIC1 expression. The mouse xenograft models of CRC confirmed that FBXW11 knockdown impeded colorectal tumor growth and liver metastasis in vivo. In summary, our study identified FBXW11 as an oncogenic factor that contributed to stem-cell-like properties and liver metastasis in CRC via regulating HIC1-mediated SIRT1 expression. These results provide a rationale for the development of FBXW11-targeting drugs for CRC patients.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

