Genomic comparison between cerebrospinal fluid and primary tumor revealed the genetic events associated with brain metastasis in lung adenocarcinoma
- PMID: 34642306
- PMCID: PMC8511004
- DOI: 10.1038/s41419-021-04223-4
Genomic comparison between cerebrospinal fluid and primary tumor revealed the genetic events associated with brain metastasis in lung adenocarcinoma
Abstract
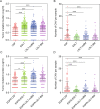
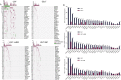
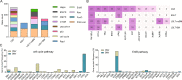
Lung adenocarcinoma (LUAD) is most common pathological type of lung cancer. LUAD with brain metastases (BMs) usually have poor prognosis. To identify the potential genetic factors associated with BM, a genomic comparison for BM cerebrospinal fluid (CSF) and primary lung tumor samples obtained from 1082 early- and late-stage LUAD patients was performed. We found that single nucleotide variation (SNV) of EGFR was highly enriched in CSF (87% of samples). Compared with the other primary lung tissues, copy number gain of EGFR (27%), CDK4 (11%), PMS2 (11%), MET (10%), IL7R (8%), RICTOR (7%), FLT4 (5%), and FGFR4 (4%), and copy number loss of CDKN2A (28%) and CDKN2B (18%) were remarkably more frequent in CSF samples. CSF had significantly lower tumor mutation burden (TMB) level but more abundant copy number variant. It was also found that the relationships among co-occurrent and mutually exclusive genes were dynamically changing with LUAD development. Additionally, CSF (97% of samples) harbored more abundant targeted drugs related driver and fusion genes. The signature 15 associated with defective DNA mismatch repair (dMMR) was only identified in the CSF group. Cancer associated pathway analysis further revealed that ErbB (95%) and cell cycle (84%) were unique pathways in CSF samples. The tumor evolution analysis showed that CSF carried significantly fewer clusters, but subclonal proportion of EGFR was remarkably increased with tumor progression. Collectively, CSF sequencing showed unique genomic characteristics and the intense copy number instability associated with cell cycle disorder and dMMR might be the crucial genetic factors in BM of LUAD.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Skřičková J, Kadlec B, Venclíček O, Merta Z. Lung cancer. Cas Lek Ceskych. 2018;157:226–36. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

