The genomic landscape of Mexican Indigenous populations brings insights into the peopling of the Americas
- PMID: 34642312
- PMCID: PMC8511047
- DOI: 10.1038/s41467-021-26188-w
The genomic landscape of Mexican Indigenous populations brings insights into the peopling of the Americas
Abstract
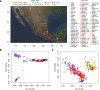
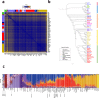
The genetic makeup of Indigenous populations inhabiting Mexico has been strongly influenced by geography and demographic history. Here, we perform a genome-wide analysis of 716 newly genotyped individuals from 60 of the 68 recognized ethnic groups in Mexico. We show that the genetic structure of these populations is strongly influenced by geography, and our demographic reconstructions suggest a decline in the population size of all tested populations in the last 15-30 generations. We find evidence that Aridoamerican and Mesoamerican populations diverged roughly 4-9.9 ka, around the time when sedentary farming started in Mesoamerica. Comparisons with ancient genomes indicate that the Upward Sun River 1 (USR1) individual is an outgroup to Mexican/South American Indigenous populations, whereas Anzick-1 was more closely related to Mesoamerican/South American populations than to those from Aridoamerica, showing an even more complex history of divergence than recognized so far.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Dixon EJ. Human colonization of the Americas: timing, technology and process. Quat. Sci. Rev. 2001;20:277–299. doi: 10.1016/S0277-3791(00)00116-5. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

