Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism
- PMID: 34642330
- PMCID: PMC8511155
- DOI: 10.1038/s41467-021-26095-0
Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism
Abstract
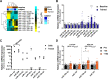
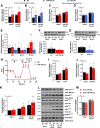
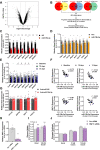
Skeletal muscle is a highly adaptable tissue and remodels in response to exercise training. Using short RNA sequencing, we determine the miRNA profile of skeletal muscle from healthy male volunteers before and after a 14-day aerobic exercise training regime. Among the exercise training-responsive miRNAs identified, miR-19b-3p was selected for further validation. Overexpression of miR-19b-3p in human skeletal muscle cells increases insulin signaling, glucose uptake, and maximal oxygen consumption, recapitulating the adaptive response to aerobic exercise training. Overexpression of miR-19b-3p in mouse flexor digitorum brevis muscle enhances contraction-induced glucose uptake, indicating that miR-19b-3p exerts control on exercise training-induced adaptations in skeletal muscle. Potential targets of miR-19b-3p that are reduced after aerobic exercise training include KIF13A, MAPK6, RNF11, and VPS37A. Amongst these, RNF11 silencing potentiates glucose uptake in human skeletal muscle cells. Collectively, we identify miR-19b-3p as an aerobic exercise training-induced miRNA that regulates skeletal muscle glucose metabolism.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

