Kincore: a web resource for structural classification of protein kinases and their inhibitors
- PMID: 34643709
- PMCID: PMC8728253
- DOI: 10.1093/nar/gkab920
Kincore: a web resource for structural classification of protein kinases and their inhibitors
Abstract
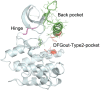
The active form of kinases is shared across different family members, as are several commonly observed inactive forms. We previously performed a clustering of the conformation of the activation loop of all protein kinase structures in the Protein Data Bank (PDB) into eight classes based on the dihedral angles that place the Phe side chain of the DFG motif at the N-terminus of the activation loop. Our clusters are strongly associated with the placement of the activation loop, the C-helix, and other structural elements of kinases. We present Kincore, a web resource providing access to our conformational assignments for kinase structures in the PDB. While other available databases provide conformational states or drug type but not both, KinCore includes the conformational state and the inhibitor type (Type 1, 1.5, 2, 3, allosteric) for each kinase chain. The user can query and browse the database using these attributes or determine the conformational labels of a kinase structure using the web server or a standalone program. The database and labeled structure files can be downloaded from the server. Kincore will help in understanding the conformational dynamics of these proteins and guide development of inhibitors targeting specific states. Kincore is available at http://dunbrack.fccc.edu/kincore.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Defining a new nomenclature for the structures of active and inactive kinases.Proc Natl Acad Sci U S A. 2019 Apr 2;116(14):6818-6827. doi: 10.1073/pnas.1814279116. Epub 2019 Mar 13. Proc Natl Acad Sci U S A. 2019. PMID: 30867294 Free PMC article.
-
Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated type II inhibitors.J Med Chem. 2015 Jan 8;58(1):466-79. doi: 10.1021/jm501603h. Epub 2014 Dec 12. J Med Chem. 2015. PMID: 25478866 Free PMC article.
-
AlphaFold2 models of the active form of all 437 catalytically competent human protein kinase domains.bioRxiv [Preprint]. 2023 Sep 3:2023.07.21.550125. doi: 10.1101/2023.07.21.550125. bioRxiv. 2023. PMID: 37547017 Free PMC article. Preprint.
-
The ABC of protein kinase conformations.Biochim Biophys Acta. 2015 Oct;1854(10 Pt B):1555-66. doi: 10.1016/j.bbapap.2015.03.009. Epub 2015 Apr 1. Biochim Biophys Acta. 2015. PMID: 25839999 Review.
-
Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes.Pharmacol Res. 2016 Jan;103:26-48. doi: 10.1016/j.phrs.2015.10.021. Epub 2015 Oct 31. Pharmacol Res. 2016. PMID: 26529477 Review.
Cited by
-
Dynamic equilibria in protein kinases.Curr Opin Struct Biol. 2021 Dec;71:215-222. doi: 10.1016/j.sbi.2021.07.006. Epub 2021 Aug 20. Curr Opin Struct Biol. 2021. PMID: 34425481 Free PMC article. Review.
-
High-throughput prediction of protein conformational distributions with subsampled AlphaFold2.Nat Commun. 2024 Mar 27;15(1):2464. doi: 10.1038/s41467-024-46715-9. Nat Commun. 2024. PMID: 38538622 Free PMC article.
-
Can Deep Learning Blind Docking Methods be Used to Predict Allosteric Compounds?J Chem Inf Model. 2025 Apr 14;65(7):3737-3748. doi: 10.1021/acs.jcim.5c00331. Epub 2025 Apr 1. J Chem Inf Model. 2025. PMID: 40167386 Free PMC article.
-
Dual-action kinase inhibitors influence p38α MAP kinase dephosphorylation.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2415150122. doi: 10.1073/pnas.2415150122. Epub 2024 Dec 31. Proc Natl Acad Sci U S A. 2025. PMID: 39739785 Free PMC article.
-
HProteome-BSite: predicted binding sites and ligands in human 3D proteome.Nucleic Acids Res. 2023 Jan 6;51(D1):D403-D408. doi: 10.1093/nar/gkac873. Nucleic Acids Res. 2023. PMID: 36243970 Free PMC article.
References
-
- Zhang J., Yang P.L., Gray N.S.. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009; 9:28–39. - PubMed
-
- Ferguson F.M., Gray N.S.. Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 2018; 17:353–377. - PubMed
-
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S.. The protein kinase complement of the human genome. Science. 2002; 298:1912–1934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

