Prevalence and Whole-Genome Sequence-Based Analysis of Shiga Toxin-Producing Escherichia coli Isolates from the Recto-Anal Junction of Slaughter-Age Irish Sheep
- PMID: 34644161
- PMCID: PMC8612287
- DOI: 10.1128/AEM.01384-21
Prevalence and Whole-Genome Sequence-Based Analysis of Shiga Toxin-Producing Escherichia coli Isolates from the Recto-Anal Junction of Slaughter-Age Irish Sheep
Abstract
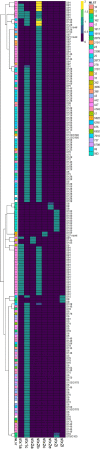
Shiga toxin-producing Escherichia coli (STEC) organisms are a diverse group of pathogenic bacteria capable of causing serious human illness, and serogroups O157 and O26 are frequently implicated in human disease. Ruminant hosts are the primary STEC reservoir, and small ruminants are important contributors to STEC transmission. This study investigated the prevalence, serotypes, and shedding dynamics of STEC, including the supershedding of serogroups O157 and O26, in Irish sheep. Recto-anal mucosal swab samples (n = 840) were collected over 24 months from two ovine slaughtering facilities. Samples were plated on selective agars and were quantitatively and qualitatively assessed via real-time PCR (RT-PCR) for Shiga toxin prevalence and serogroup. A subset of STEC isolates (n = 199) were selected for whole-genome sequencing and analyzed in silico. In total, 704/840 (83.8%) swab samples were Shiga toxin positive following RT-PCR screening, and 363/704 (51.6%) animals were subsequently culture positive for STEC. Five animals were shedding STEC O157, and three of these were identified as supershedders. No STEC O26 was isolated. Post hoc statistical analysis showed that younger animals are more likely to harbor STEC and that STEC carriage is most prevalent during the summer months. Following sequencing, 178/199 genomes were confirmed as STEC. Thirty-five different serotypes were identified, 15 of which were not yet reported for sheep. Serotype O91:H14 was the most frequently reported. Eight Shiga toxin gene variants were reported, two stx1 and six stx2, and three novel Shiga-toxin subunit combinations were observed. Variant stx1c was the most prevalent, while many strains also harbored stx2b. IMPORTANCE Shiga toxin-producing Escherichia coli (STEC) bacteria are foodborne, zoonotic pathogens of significant public health concern. All STEC organisms harbor stx, a critical virulence determinant, but it is not expressed in most serotypes. Sheep shed the pathogen via fecal excretion and are increasingly recognized as important contributors to the dissemination of STEC. In this study, we have found that there is high prevalence of STEC circulating within sheep and that prevalence is related to animal age and seasonality. Further, sheep harbor a variety of non-O157 STEC, whose prevalence and contribution to human disease have been underinvestigated for many years. A variety of Stx variants were also observed, some of which are of high clinical importance.
Keywords: Shiga toxin-producing Escherichia coli; non-O157 STEC; sheep; supershedding; whole-genome sequencing.
Figures
References
-
- Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, Yeung DH, Kirk MD. 2014. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis 11:447–455. 10.1089/fpd.2013.1704. - DOI - PMC - PubMed
-
- Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O’Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. 10.1128/JCM.00860-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

