Liquid Biopsy in Gastric Cancer: Analysis of Somatic Cancer Tissue Mutations in Plasma Cell-Free DNA for Predicting Disease State and Patient Survival
- PMID: 34644276
- PMCID: PMC8462609
- DOI: 10.14309/ctg.0000000000000403
Liquid Biopsy in Gastric Cancer: Analysis of Somatic Cancer Tissue Mutations in Plasma Cell-Free DNA for Predicting Disease State and Patient Survival
Abstract
Introduction: Gastric cancer (GC) diagnosis in late stages and high mortality rates are the main issues that require new noninvasive molecular tools. We aimed to assess somatic mutational profiles in GC tissue and plasma cell-free DNA (cfDNA), evaluate their concordance rate, and analyze the role of multilayer molecular profiling to predict disease state and prognosis.
Methods: Treatment-naive GC patient group (n = 29) was selected. Whole exome sequencing (WES) of GC tissue was performed, and a unique 38-gene panel for deep targeted sequencing of plasma cfDNA was developed. Oncoproteins were measured by enzyme-linked immunosorbent assay, and other variables such as tumor mutational burden and microsatellite instability were evaluated using WES data.
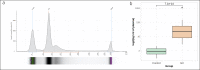
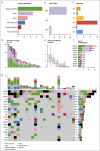
Results: The yield of cfDNA was increased 43.6-fold; the integrity of fragments was decreased in GC compared with controls. WES analysis of cancerous tissue and plasma cfDNA (targeted sequencing) mutational profiles revealed 47.8% concordance. The increased quantity of GC tissue-derived alterations detected in cfDNA was associated with worse patients' survival. Analysis of importance of multilayer variables and receiver operating characteristic curve showed that combination of 2 analytes: (i) quantity of tissue matching alterations and (ii) presence of any somatic alteration in plasma cfDNA resulted in area under curve 0.744 when discriminating patients with or without distant metastasis. Furthermore, cfDNA sequence alterations derived from tumor tissue were detected in patients who had even relatively small GC tumors (T1-T2).
Discussion: Our results indicate that quantitative and qualitative cfDNA mutational profile analysis is a promising tool for evaluating GC disease status or poorer prognosis.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of The American College of Gastroenterology.
Conflict of interest statement
Figures




References
-
- Bray F, Ferlay J, Soerjomataram I, et al. . Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. - PubMed
-
- Rajilic-Stojanovic M, Figueiredo C, Smet A, et al. Systematic review: Gastric microbiota in health and disease. Aliment Pharmacol Ther 2020;51:582–602. - PubMed
-
- Bornschein J, Leja M, Kupcinskas J, et al. . Molecular diagnostics in gastric cancer. Front Biosci 2014;19:312–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

