IL-33-induced metabolic reprogramming controls the differentiation of alternatively activated macrophages and the resolution of inflammation
- PMID: 34644537
- PMCID: PMC7617137
- DOI: 10.1016/j.immuni.2021.09.010
IL-33-induced metabolic reprogramming controls the differentiation of alternatively activated macrophages and the resolution of inflammation
Abstract
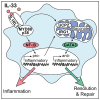
Alternatively activated macrophages (AAMs) contribute to the resolution of inflammation and tissue repair. However, molecular pathways that govern their differentiation have remained incompletely understood. Here, we show that uncoupling protein-2-mediated mitochondrial reprogramming and the transcription factor GATA3 specifically controlled the differentiation of pro-resolving AAMs in response to the alarmin IL-33. In macrophages, IL-33 sequentially triggered early expression of pro-inflammatory genes and subsequent differentiation into AAMs. Global analysis of underlying signaling events revealed that IL-33 induced a rapid metabolic rewiring of macrophages that involved uncoupling of the respiratory chain and increased production of the metabolite itaconate, which subsequently triggered a GATA3-mediated AAM polarization. Conditional deletion of GATA3 in mononuclear phagocytes accordingly abrogated IL-33-induced differentiation of AAMs and tissue repair upon muscle injury. Our data thus identify an IL-4-independent and GATA3-dependent pathway in mononuclear phagocytes that results from mitochondrial rewiring and controls macrophage plasticity and the resolution of inflammation.
Keywords: GATA3; UCP2; alternatively activated macrophage; interleukin-33; itaconate; mitochondrial rewiring; resolution of inlammation; uncoupling.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests
Figures






References
-
- Chazaud B. Inflammation and skeletal muscle regeneration: leave it to the macrophages! Trends Immunol. 2020;41:481–492. - PubMed
-
- Coakley G, Harris NL. Interactions between macrophages and helminths. Parasite Immunol. 2020;42:e12717. - PubMed
-
- Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

